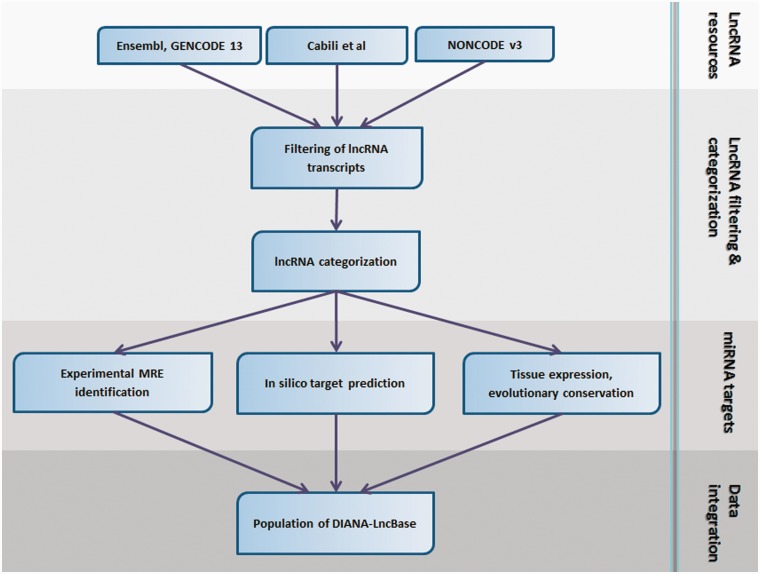
Figure 1.
DIANA-LncBase analysis pipeline. The best available lncRNA resources have been collected for human and mouse species. Transcripts shorter than 200 nt as well as transcripts presenting high similarity (>90% overlap) have been removed. lncRNAs have been subsequently categorized in sense, antisense, bidirectional and intergenic with the use of a protein-coding reference set consisting of UCSC and Ensembl genes. Additionally, MREs on lncRNAs have been experimentally verified with the use of high-throughput HITS/PAR-CLIP data and in silico predicted with a state-of-the-art algorithm, DIANA-microT-CDS. Integration of miRNA targets, as well as other lncRNA/miRNA-related information, such as transcripts tissue expression and MREs evolutionary conservation, has been the final step for DIANA-LncBase population.