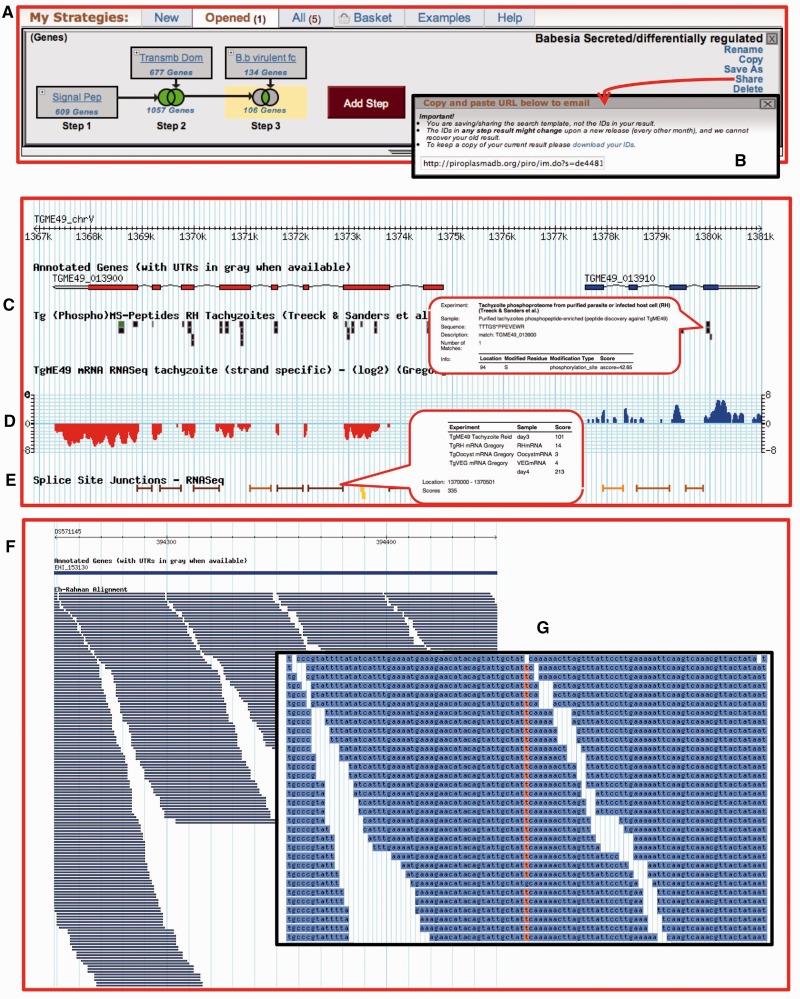
Figure 1.
Screen shots of a search strategy in PiroplasmaDB and GBrowse representing HTS (C–E from ToxoDB and F and G from AmoebaDB) (A) A three-step search strategy combining genes with predicted signal peptides, transmembrane domains and microarray expression data. (B) Search strategies may be saved and shared with others using a uniquely generated URL. (C) Peptides from mass spec experiments are mapped to genes and displayed graphically. Mousing over the graphics provides additional information, such as the peptide sequence and any posttranslational modifications. In this image, peptides are from a phophoproteomic experiment. (D) A track representing strand-specific RNA-seq data. Blue indicates reads mapping to the forward strand, whereas red represents those mapping to the reverse strand. (E) Unified splice junction track representing intron-spanning RNA-seq reads from all experiments in the database. (F) A 2 kb region with alignment of DNA sequencing reads to the genome. (G) Zooming in to 100 bp displays the actual sequence allowing data inspection. Highlighted nucleotides represent SNPs.