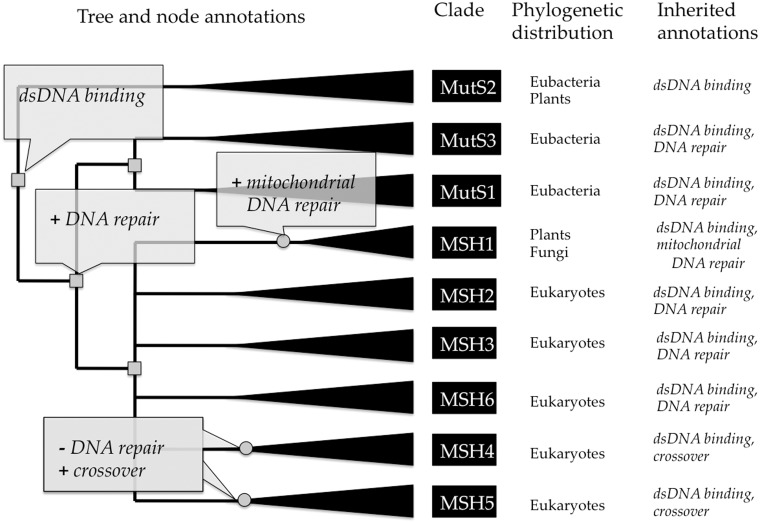
Figure 5.
Example tree annotations of gene function, for the MutS homolog family (PTHR11361). The tree is annotated with functional ‘gains’ (indicated by plus) and ‘losses’ (indicated by minus). Inferred gene duplications are indicated by small gray squares, and annotated speciation events by small gray circles. Based on the distribution of experimental GO annotations from the literature for genes in model organisms (not shown here), the curator has inferred that the common ancestor of the family had the molecular function ‘double-stranded DNA binding’. Prior to the common ancestor of mutS (bacteria) and the mutS homologs (MSH’s in eukaryotes), one duplicate gained a function in ‘DNA repair’ (biological process). This function was preserved in most descendant homologs, but was lost prior to the radiation of MSH4 and MSH5. In the MSH1 lineage, a specific function in ‘mitochondrial DNA repair’ was gained (likely through the endosymbiotic origin of mitochondria). Extant sequences (leaves of the tree) are annotated by inheritance from the ancestral node annotations. Note that all ancestral annotations are inherited unless they are lost in a more recent ancestor, e.g. MSH4 and MSH5 do not inherit DNA repair even though they have an ancestor that likely participated in DNA repair. Full annotations for this family are available at http://www.pantree.org/tree/family.jsp?accession=PTHR11361.