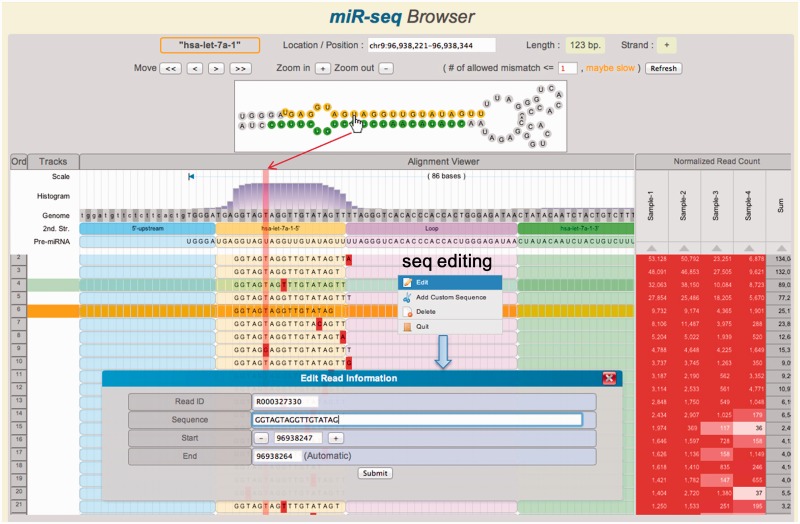
Figure 2.
Main features of the miR-seq Browser. At the top panel, the hairpin structure of miRNA precursor is shown. The aligned short reads are shown together with secondary structure and read depth information in the track. By mouseover (hand icon) on a nucleotide, the corresponding columns are highlighted in vertical pink shadow. The reads can be sorted by the read count of each sample or the total sum on the right panel. Note that several read sequences show 3′-end modifications. Histogram shows the read depth at each position. Mismatched nucleotides are highlighted in red. Sequence editor window is opened by right-click.