Abstract
Recent studies have unravelled the diversity of sponge-associated bacteria that may play essential roles in sponge health and metabolism. Nevertheless, our understanding of this microbiota remains limited to a few host species found in restricted geographical localities, and the extent to which the sponge host determines the composition of its own microbiome remains a matter of debate. We address bacterial abundance and diversity of two temperate marine sponges belonging to the Irciniidae family - Sarcotragus spinosulus and Ircinia variabilis – in the Northeast Atlantic. Epifluorescence microscopy revealed that S. spinosulus hosted significantly more prokaryotic cells than I. variabilis and that prokaryotic abundance in both species was about 4 orders of magnitude higher than in seawater. Polymerase chain reaction-denaturing gradient gel electrophoresis (PCR-DGGE) profiles of S. spinosulus and I. variabilis differed markedly from each other – with higher number of ribotypes observed in S. spinosulus – and from those of seawater. Four PCR-DGGE bands, two specific to S. spinosulus, one specific to I. variabilis, and one present in both sponge species, affiliated with an uncultured sponge-specific phylogenetic cluster in the order Acidimicrobiales (Actinobacteria). Two PCR-DGGE bands present exclusively in S. spinosulus fingerprints affiliated with one sponge-specific phylogenetic cluster in the phylum Chloroflexi and with sponge-derived sequences in the order Chromatiales (Gammaproteobacteria), respectively. One Alphaproteobacteria band specific to S. spinosulus was placed in an uncultured sponge-specific phylogenetic cluster with a close relationship to the genus Rhodovulum. Our results confirm the hypothesized host-specific composition of bacterial communities between phylogenetically and spatially close sponge species in the Irciniidae family, with S. spinosulus displaying higher bacterial community diversity and distinctiveness than I. variabilis. These findings suggest a pivotal host-driven effect on the shape of the marine sponge microbiome, bearing implications to our current understanding of the distribution of microbial genetic resources in the marine realm.
Introduction
Marine sponges have been the focus of increasing microbiology research interest mainly because of their symbiotic association with abundant and diverse bacteria and production of biologically active secondary metabolites [1], [2]. For so-called High Microbial Abundance (HMA) sponges, it has been shown that up to 38% of sponge wet weight is composed of bacterial cells [3], and that such bacterial abundance surpasses that of seawater by 2 to 4 orders of magnitude [1], [4]–[6]. It has been suggested that HMA sponges harbour several bacteria involved in the production of secondary metabolites, which might, for example, improve protection against predation of the sponge host [1]. The synthesis of bioactive compounds derived from sponge-microbe associations has already been reported for 26 of the 92 families in the Demospongia [7], the most diversified class of the phylum Porifera. Currently, the use of high-throughput sequencing technology is extending our knowledge of microbial diversity in marine sponges, with more than 25 bacterial phyla detected in sponges by this means [8], [9]. Taken together, these features foreshadow marine sponge holobiomes as valuable reservoirs of microbial genetic and metabolic diversity of potential use in biotechnology.
Despite such remarkable advances, current understanding of symbiont community structure in marine sponges remains restricted to certain regions and host species [1]. This holds true for species within the family Irciniidae (Demospongiae, Dictyoceratida), from which the majority of surveys undertaken so far have been limited to tropical latitudes and to the species Ircinia felix, I. strobilina, and I. ramosa [8], [10]–[17]. Electron microscopy analyses unveiled abundant and diverse bacterial morphotypes in I. felix [10]–[12], whereas five [15] and seven [17] bacterial phyla were revealed in association with I. strobilina by cloning-and-sequencing of 16S rRNAgene fragments. By means of high-throughput sequencing, sixteen phyla and 1199 bacterial operational taxonomic units (OTUs) at 95% sequence similarity were found in association with I. ramosa [8], highlighting the complexity of the Ircinia-associated “bacteriome”. The detection of Acidobacteria, Alpha- and Gammaproteobacteria in adult, larva and juvenile samples of I. felix [11] supports the hypothesis that a portion of this microbiota might be vertically transmitted throughout successive host generations.
Conversely, the microbial ecology of temperate irciniids remains underexplored. Only recently a study first approached the diversity of bacterial communities in Mediterranean Ircinia spp. - namely I. variabilis, I. fasciculata and I. oros - revealing eight bacterial phyla across these hosts and species-specific OTUs [18]. Because of their global distribution, encompassing both tropical and temperate species, Irciniidae sponges constitute a valuable taxon for the study of the ecology and evolution of symbiotic relationships. In addition, a great variety of cytotoxic compounds has been retrieved from Irciniidae species, which indicates they are potentially of high biotechnological importance. [19]–[24]. Furthermore, two studies performed with the temperate I. muscarum and I. variabilis described the production of cyclic peptides by cultivated bacteria [25], [26], whereas psymberin – which resembled the pederin family of polyketides – was recovered from Psammocinia sp. and shown to have a bacterial symbiont origin [19], [27]. In this context, addressing microbial diversity and distribution in widespread and chemically complex marine sponges is not only relevant to the study of symbiosis and co-evolutionary relationships, but also bears importance to our understanding of the extent of marine genetic and metabolic resources.
In light of the recent evidence for divergent bacterial communities across different sponge species or even specimens [9], [28], a feature that has also been observed for other eukaryotes that support complex bacterial consortia [29]–[31], this study uses a stringent experimental design to test the hypothesis of host-specific assemblages of dominant symbionts in marine sponges. To this end, we address bacterial abundance and diversity in the temperate marine sponges Sarcotragus spinosulus Schmidt, 1862 and Ircinia variabilis Schmidt, 1862 (Demospongiae, Dictyoceratida, Irciniidae), two closely related species that co-exist in spatial proximity at the coast of the Algarve (southern Portugal), a region with a Mediterranean climate located in the Northeast Atlantic. We use the 16S rRNA gene as a phylogenetic marker in polymerase chain reaction – denaturing gradient gel electrophoresis (PCR-DGGE) analyses of the domain Bacteria, the phylum Actinobacteria and the class Alphaproteobacteria in these hosts, thus allowing the inspection of bacterial community structures across different taxonomic ranks with concomitant focus on abundant and biotechnologically relevant sponge-associated microorganisms [7], [32], [33]. Phylogenetic analysis of dominant bacterial populations (i.e. PCR-DGGE bands) consistently and specifically found in association with these species is performed, and their status as “sponge-specific bacterial clusters” [34] is verified. We also determine the degree of dissimilarity between sponge-associated bacterial communities and that of their neighbouring bacterioplankton. To assure accurate identification of the target sponges, we infer host phylogenies based on cytochrome oxidase gene sequence relationships. This is the first study of bacterial community structure and diversity in North Atlantic Irciniidae.
Results
Sponge Identification
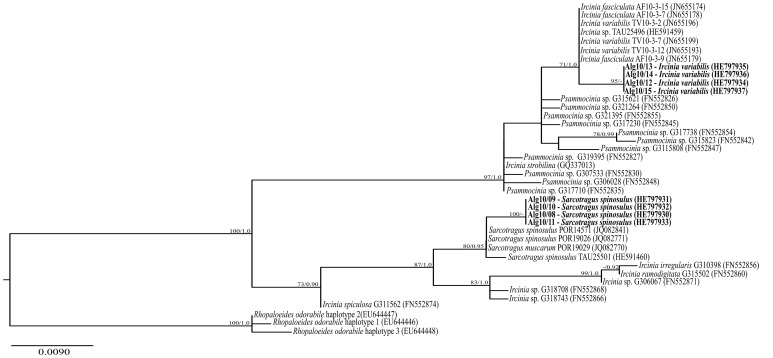
Sponge specimens (Fig. S1) were identified as Sarcotragus spinosulus and Ircinia variabilis based on macro- and microscopic analyses using morphological criteria. Analysis of 636 bp-long sequences of the subunit I of the mitochondrial cytochrome C oxidase (CO1) gene obtained for all specimens (accession numbers HE797930 to HE797937) showed no intraspecific variation among our sequences of I. variabilis or S. spinosulus, whereas a 4.7% genetic distance (p-distance) was found between our sequences of these two species. Genetic distances between our S. spinosulus sequences and those available on NCBI GenBank ranged from 0 to 0.6%, whereas for I. variabilis a distance of 0.5% was observed between our sequences and those of I. variabilis/fasciculata collected in the Northwestern Mediterranean. Phylogenetic reconstructions based both on Maximum Likelihood and Bayesian inference confirmed the identification of our sponge specimens. Indeed, I. variabilis and S. spinosulus CO1 sequences sampled in this study formed well-supported clades with CO1 sequences from Ircinia spp. and Sarcotragus spp. retrieved elsewhere (Fig. 1).
Figure 1. Phylogenetic inference of the Irciniidae family based on the cytochrome oxidase gene, subunit 1.
The Maximum Likelihood tree (-ln likelihood: 1383.921591) is shown, with sequences retrieved in this study highlighted in bold. Numbers at tree nodes are bootstrap values and posterior probabilities calculated in Maximum Likelihood and MCMC Bayesian analyses, respectively, and values above 70/0.95 are shown.
Counting of Heterotrophic Culturable Bacteria
The colony forming units (CFU) counts of heterotrophic bacteria on marine agar revealed no significant difference (p>0.05) between sponges species, with 3.21±2.03×106 CFU and 1.63±0.61×106 CFU g−1 of fresh sponge for S. spinosulus and I. variabilis, respectively.
Epifluorescence Microscopy
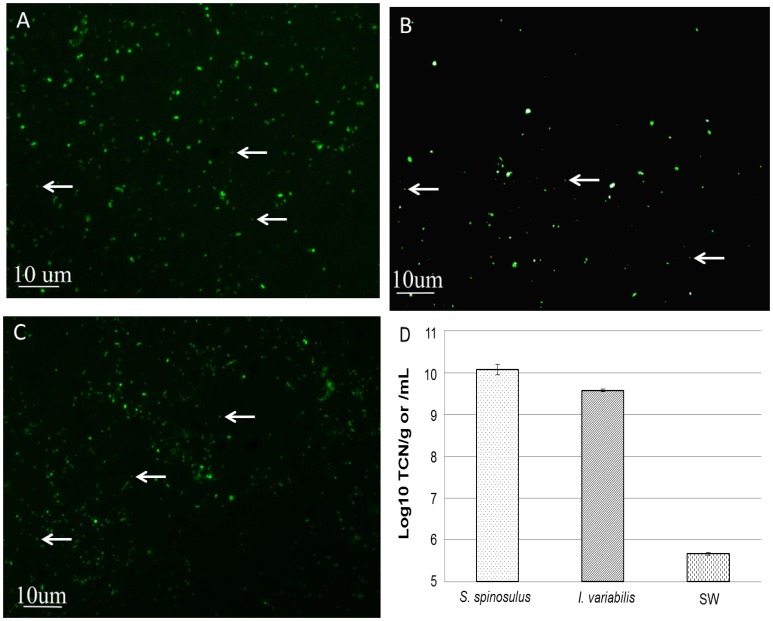
Analyses showed that S. spinosulus harboured significantly (p<0.05) higher number of prokaryotic cells (average of 1.37×1010 cells g−1 of fresh sponge), as surveyed in this study, when compared to I. variabilis (average of 3.81×109 cells g−1 of fresh sponge). The abundance of prokaryotic cells in surrounding seawater (average of 4.63×105 cells mL−1) was significantly (p<0.05) lower than in both sponge species (Fig. 2).
Figure 2. Epifluorescence counts.
Microscopy pictures taken from S. spinosulus (A), I. variabilis (B) and Seawater (C) are shown. Arrows exemplify counted bacterial cells. Values in panel (D) are expressed as means ± standard errors of log-transformed total cell numbers (TCN).
PCR-DGGE Analysis of Bacterial Communities
(i) Bacteria PCR-DGGE profiles
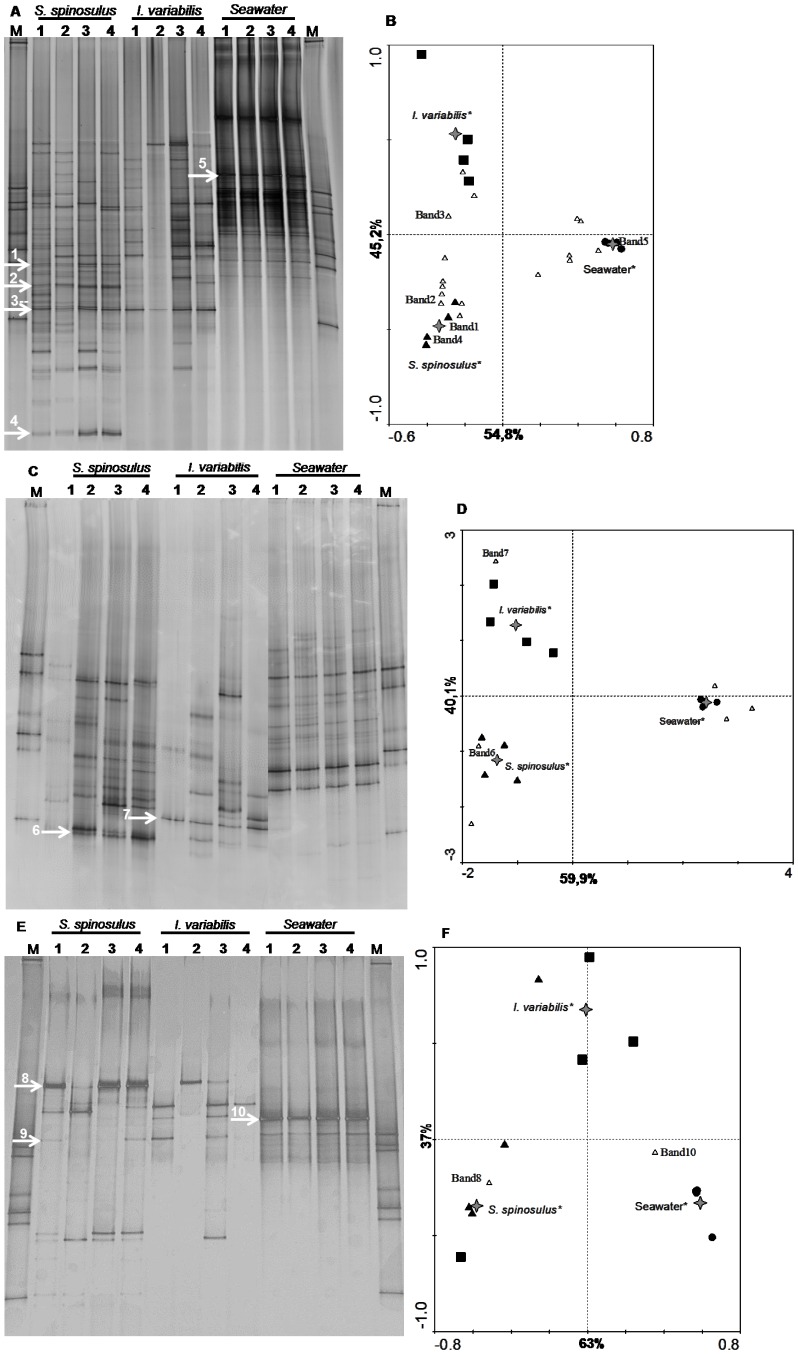
The bacterial PCR-DGGE profiles of S. spinosulus were characterized by 5 dominant bands and a large number of fainter bands (16 to 30) whereas those of I. variabilis comprised 1 dominant band in addition to 5 to 26 fainter bands (Fig. 3a, Table S1). Seawater DGGE profiles showed 7 dominant bands and a large number of fainter bands (28 to 30). While the similarity within seawater and S. spinosulus replicates was high, profiles of I. variabilis specimens displayed large within-replicate heterogeneity (Fig. 3a). Clearly contrasting profiles were observed between seawater and sponge samples, and between both sponge species. Indeed, the UPGMA cluster analysis (Fig. S2a) revealed two main groups, one formed exclusively by all sponge specimens and other containing only seawater samples. These two groups shared less than 10% similarity whereas S. spinosulus and I. variabilis PCR-DGGE profiles shared around 20% similarity. Ordination via canonical correspondence analysis (CCA) of the DGGE band data and environment variables revealed that sponge species and seawater significantly influenced band variation in the DGGE profiles (p<0.05, Fig. 3b). The horizontal axis of the diagram, which accounted for 54.8% of the dependent (i.e. DGGE bands) – independent (i.e. sample classes) variables correlations, mainly distinguished the sponges S. spinosulus and I. variabilis from seawater (Fig. 3b). The vertical axis grouped replicates from sponge S. spinosulus clearly apart from those observed in I. variabilis.
Figure 3. PCR-DGGE fingerprints and canonical analyses.
PCR-DGGE 16S rRNA gene fingerprints of S. spinosulus, I. variabilis and seawater DNA samples generated with “total-community” bacterial primers (A) and specific primer systems for Actinobacteria (C) and Alphaproteobacteria (E). The arrows indicate bands that were excised from DG-gels and sequenced. Corresponding ordination biplots of PCR-DGGE fingerprints and qualitative environmental variables are shown in panels B, D, F. Symbols: ▴ S. spinosulus, ▪ I. variabilis and • Seawater. Labels displayed on the diagram axes refer to the percentage variations of PCR-DGGE ribotypes - environment correlation accounted for the respective axis. The “star” symbols represent the centroid positions of the environmental variables in the diagram. Variables that significantly (p<0.05) influence the bacterial community composition are indicated by an asterisk.
(ii) Actinobacteria PCR-DGGE profiles
The actinobacterial PCR-DGGE profiles of S. spinosulus consisted of few (2 to 4) strong bands along with more than 4 detectable bands, while those of I. variabilis comprised 1 to 3 dominant bands along with 1 to 5 fainter bands (Fig. 3c). Significant reduction in actinobacterial diversity and richness were determined for the latter species in comparison with the former (Table S1). A large heterogeneity was observed within I. variabilis profiles. In comparison with the sponge fingerprints, the seawater PCR-DGGE profiles displayed higher diversity of bands, especially against I. variabilis profiles (Table S1), and contained 3 strong bands along with more than 6 fainter bands, and much lower within-replicate variability (Fig. 3c). Two main groups were revealed by cluster analysis, one formed exclusively by all sponge specimens and other containing only seawater samples (Fig. S2b). These two groups shared about 10% similarity. However, there was also a clear difference between the profiles of both sponge species, which shared c. 20% similarity according to cluster analysis. Ordination via CCA discriminated both sponge species and seawater across the horizontal axis of the diagram, which represented around 60% of the overall PCR-DGGE – sample correlations (Fig. 3d). All independent variables (i.e. the sample classes seawater, S. spinosulus and I. variabilis) significantly (p<0.05) affected the PCR-DGGE banding patterns (Fig. 3d).
(iii) Alphaproteobacteria PCR-DGGE profiles
The Alphaproteobacteria profiles of S. spinosulus contained 1 to 3 dominant bands, in addition to more than 7 detectable fainter bands, whereas the profiles of I. variabilis revealed 1 to 3 strong and fainter bands (Fig. 3e). Significantly greater richness, diversity and evenness were found for S. spinosulus alphaproteobacterial PCR-DGGE profiles in comparison with those of I. variabilis (Table S1). The seawater profiles showed 2 strong bands along with more than 6 fainter bands. The variability within sponge specimens and among sponge species was relatively high. Conversely, seawater samples displayed highly homogeneous profiles (Fig. 3e). Cluster analysis revealed a clear separation between seawater and sponge samples and high similarity scores for the former (Fig. S2c). The latter grouped into two further clusters in which the visible, higher degrees of within-sample variability could be numerically depicted (Fig. S2c). Sample outliers were detected, as one replicate from I. variabilis grouped with a cluster dominated by three S. spinosulus samples, and the same effect was observed for one replicate from S. spinosulus which clustered with I. variabilis specimens (Fig. 3f, S1c). Nevertheless, CCA showed that all factors significantly (p<0.05) influenced the patterns of band distribution in alphaproteobacterial PCR-DGGE profiles. Ordination via CCA revealed that 63% of total PCR-DGGE band – independent variables correlations was explained by the horizontal axis of the diagram, which mainly differentiated S. spinosulus from seawater (Fig. 3f), whereas the residual variability in the vertical axis of the diagram (37%) discriminated most I. variabilis from seawater and S. spinosulus samples (Fig. 3f).
Analysis of Sequences of Dominant and Discriminating PCR-DGGE Bands
(i) Bacteria PCR-DGGE bands
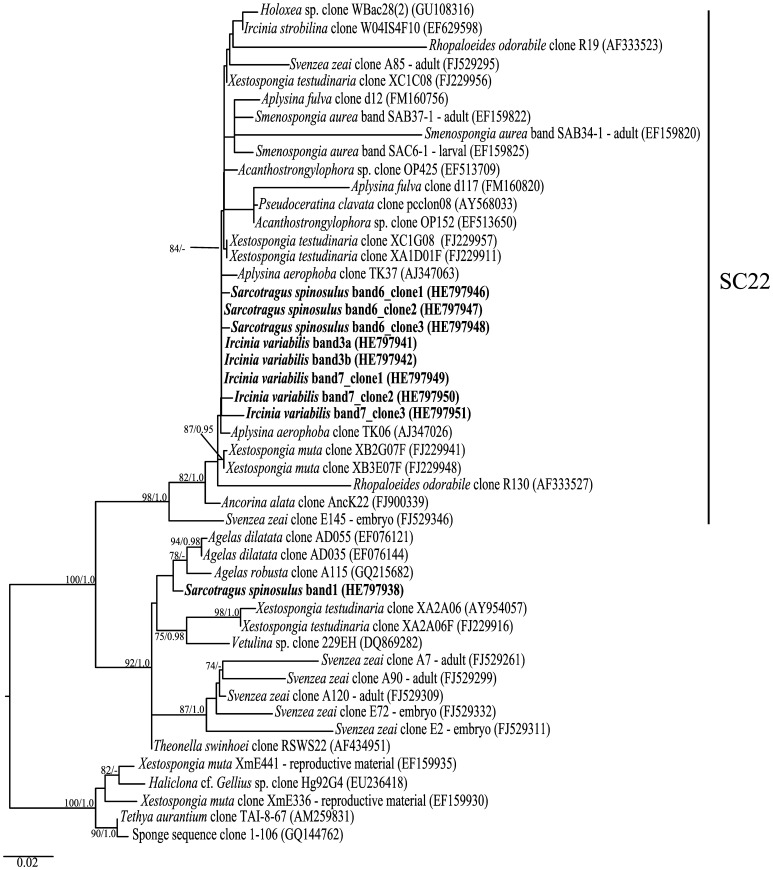
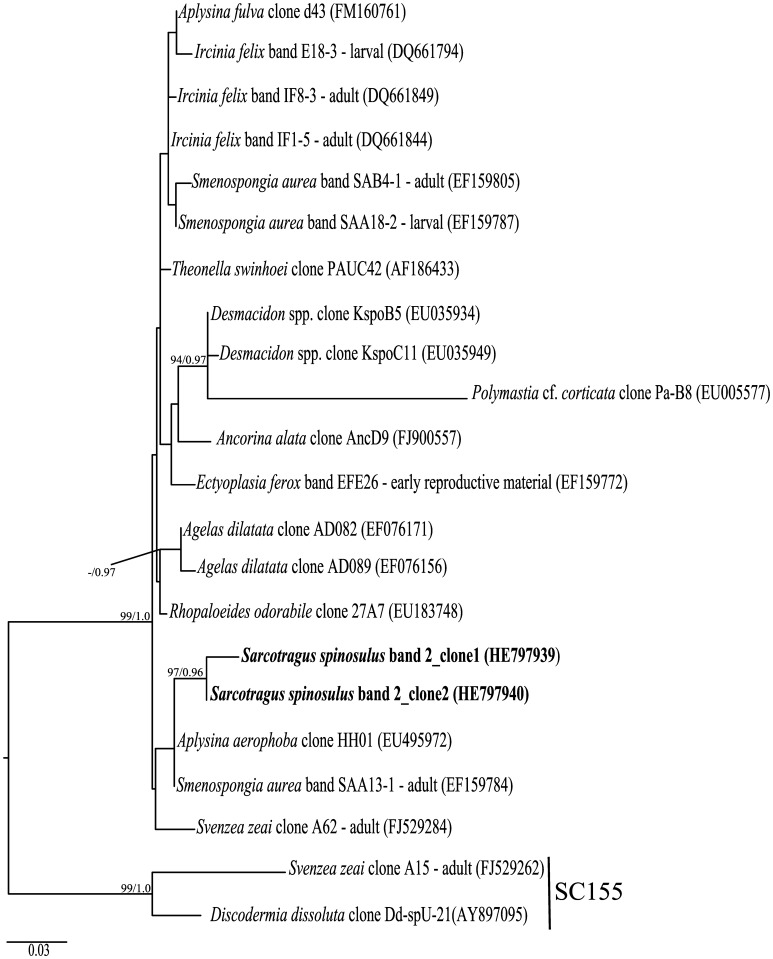
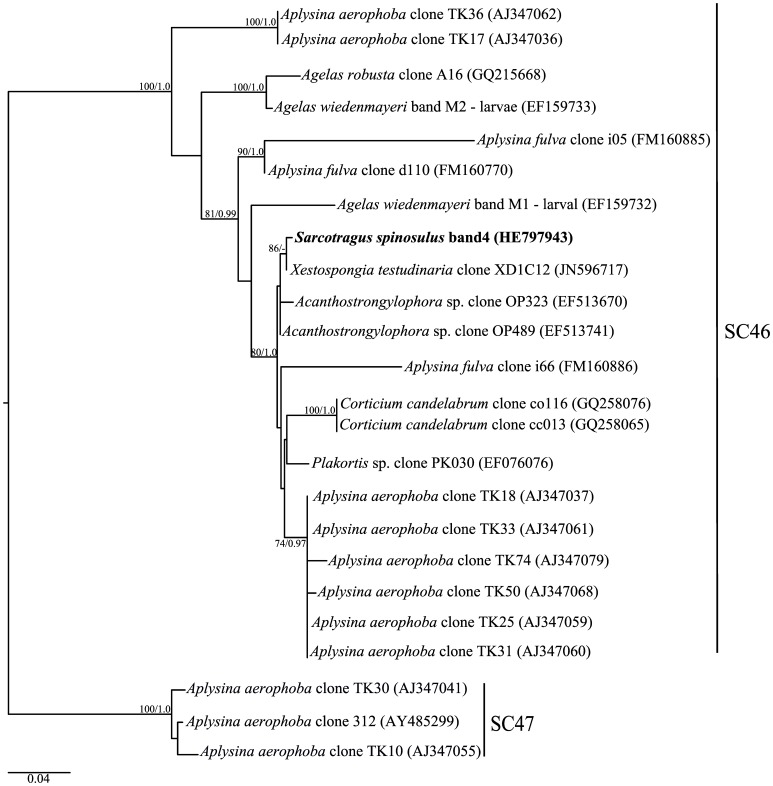
Three dominant bands labelled 1, 2 and 4 (see arrows in Fig. 3a) were exclusively found in all replicates of S. spinosulus. Bands 1 and 4 were directly sequenced whereas band 2 was subjected to cloning and sequencing. From band 1, one sequence was retrieved and affiliated with the Actinobacteria order Acidimicrobiales. The phylogenetic analysis showed that this band affiliated with an uncultured and apparently diverse lineage containing sponge-derived bacterial sequences of worldwide origin (Fig. 4). Further, two clones were sequenced from band 2 and found to be highly alike, with 5 different nucleotides between them. They were assigned to the Gammaproteobacteria order Chromatiales. These sequences belonged to a well supported Chromatiales phylogenetic clade containing uncultured bacteria retrieved exclusively from marine sponges sampled in several geographical localities (Fig. 5). From band 4, one sequence was obtained and classified in the Chloroflexi phylum. Phylogenetic analysis revealed that this sequence belonged to a sponge-specific bacterial phylogenetic cluster as determined by Simister et al. [34] (Fig. 6). A dominant band labelled 3 (Fig. 3a) was found in all sponge specimens. Two identical sequences were recovered and assigned to the Actinobacteria order Acidimicrobiales. They also affiliated with an uncultured sponge-specific lineage previously suggested by Simister et al. [34] (Fig. 4). A dominant band labelled 5 (see arrow in Fig. 3a) was exclusively found in all seawater samples. This band possessed high discriminating power, as obviated by its centroid position in the CCA diagram. Two identical sequences were obtained for this band and assigned to the Alphaproteobacteria family Rhodobacteraceae (Table 1). They belonged to a supported, uncultured bacterial phylogenetic clade containing sequences retrieved solely from seawater (data not shown).
Figure 4. Phylogenetic inference of Actinobacteria 16S rRNA genes.
The modified ARB database generated by Simister et al., [34] used long sequences (≥1200 bp) to infer the phylogeny and shorter sequences were added using the ARB parsimony interactive tool. Sequences from the sponge-specific cluster 22 (SC22) [34] along with sequences closely related to band 1 and outgroup sequences were selected for further phylogenetic analysis. The Maximum Likelihood tree (-ln likelihood: 4501,317092) is shown, with sequences retrieved in this study highlighted in bold. Numbers at tree nodes are bootstrap values and posterior probabilities calculated in Maximum Likelihood and MCMC Bayesian analyses, respectively, and values above 70/0.95 are shown.
Figure 5. Phylogenetic inference of Gammaproteobacteria 16S rRNA genes.
Tree construction procedure was as described for Figure 4, except that sequences closely related to band 2 were selected as well as sequences from SC155 [34], which were used as outgroup. The Maximum Likelihood tree is shown (-ln likelihood: 2696,593494).
Figure 6. Phylogenetic inference of Chloroflexi 16S rRNA genes.
Tree construction procedure was as described for Figure 4, except that sequences from SC46 were selected along with sequences from SC47, which were used as outgroup [34]. The Maximum Likelihood tree is shown (-ln likelihood: 3791,095).
Table 1. Closest 16S rRNA gene relatives of seawater-derived and “cosmopolitan” PCR-DGGE bands.
| Band id (Accession number) | RDP closest match1 (Accession number) | NCBI closest match2 (Percent similarity, accession number) |
| 5a (HE797944) | Uncultured Roseobacter sp. (AY627365) | Uncultured Rhodobacteraceae bacterium clone GG101008Clone5 (99%, JN591908) |
| 5b (HE797945) | Uncultured Roseobacter sp. (AY627365) | Uncultured Rhodobacteraceae bacterium clone GG101008Clone5 (99%, JN591908) |
| 9a (HE797955) | Uncultured Roseobacter sp. (AY627365) | Uncultured Rhodobacteraceae bacterium clone GG101008Clone5 (99%, JN591908) |
| 10a (HE797957) | Uncultured Roseobacter sp. (AY627365) | Uncultured Rhodobacteraceae bacterium clone GG101008Clone5 (100%, JN591908) |
| 10b (HE797958) | Uncultured Roseobacter sp. (AY627365) | Uncultured Rhodobacteraceae bacterium clone GG101008Clone5 (100%, JN591908) |
| 10c (HE797956) | Uncultured Roseobacter sp. (AY627365) | Uncultured Rhodobacteraceae bacterium clone GG101008Clone5 (100%, JN591908) |
Closest 16S rRNA gene relative using the “sequence match” tool of the Ribosomal Database Project (RDP).
Closest 16S rRNA gene relatives as determined by the blast-n search in the National Center for Biotechnology Information (NCBI) database.
(ii) Actinobacteria PCR-DGGE bands
The dominant bands labelled 6 and 7 (Fig. 3C) were recovered from three specimens of S. spinosulus and from all I. variabilis specimens, respectively. These bands were subjected to cloning and sequencing. Three clones were obtained from each band, which contained 2 and 3 dissimilar nucleotides for bands 6 and 7, respectively. All sequences were assigned to the order Acidimicrobiales. Phylogenetic analysis revealed that these sequences fell into a sponge-specific bacterial phylogenetic clade [34] from which no cultured representative has so far been registered (Fig. 4).
(iii) Alphaproteobacteria PCR-DGGE bands
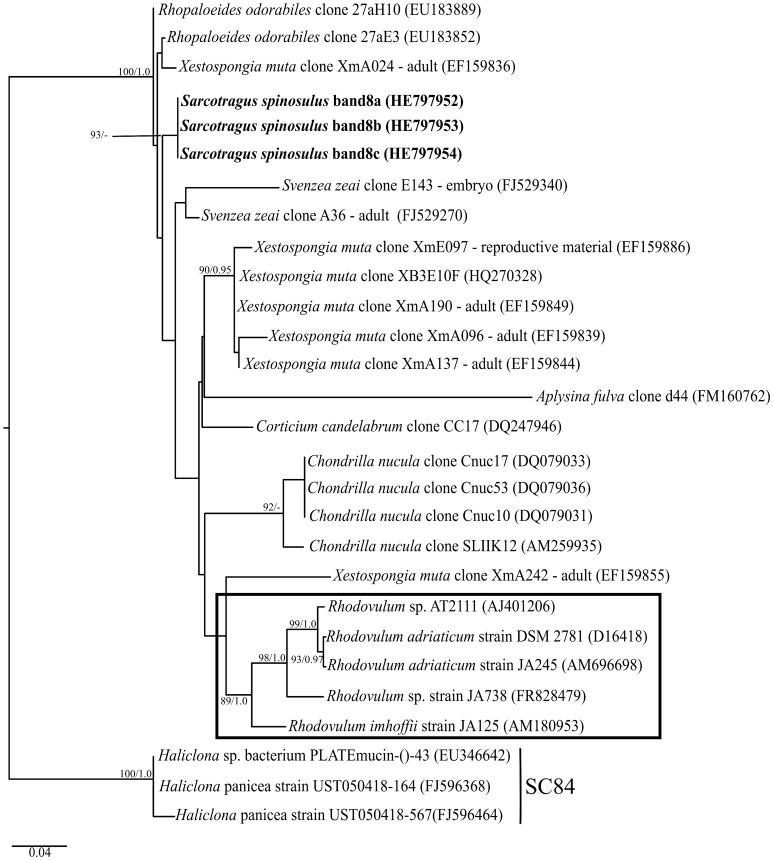
The dominant band labelled 8 (Fig. 3e) appeared in all specimens of S. spinosulus. Three identical sequences were recovered and assigned to the order Rhodobacterales. Phylogenetic reconstruction revealed that these sequences affiliated with bacterial phylotypes retrieved almost solely from marine sponges distributed worldwide. No cultured representative isolated from marine sponges has been observed in this cluster (Fig. 7). One band found almost in all samples in addition to a dominant band found exclusively in seawater samples labelled, respectively, 9 and 10 (Fig. 3e) were subjected to sequencing. One and three sequences were obtained from bands 9 and 10, respectively. They all shared high similarity at the primary sequence level (up to 3 nucleotide differences detected), belonged to a phylogenetic cluster containing several uncultured bacterial phylotypes retrieved only from seawater, and affiliated with the family Rhodobacteraceae (Table 1).
Figure 7. Phylogenetic inference of Alphaproteobacteria 16S rRNA genes.
Tree construction procedure was as described for Figure 4, except that sequences close related to band 8 were selected along with sequences from SC84 [34], which were used as outgroup. The sequences shown in a box were isolated from different environments. The Maximum Likelihood tree is shown (-ln likelihood: 3253,686594).
Discussion
This survey addresses bacterial abundance, diversity and specificity in the Atlanto-Mediterranean sponges Sarcotragus spinosulus and Ircinia variabilis (Demospongiae, Dictyoceratida, Irciniidae). These species are widely distributed along the southern Portuguese coast (http://www.marinespecies.org/porifera/). Both species were initially identified by traditional taxonomic methods. However, species within the Order Dictyoceratida to which the family Irciniidae belongs are, along with the Order Dendroceratida, known as ‘keratose’ sponges, which usually lack a suite of morphological features making their classification problematic [35], [36]. In recent years, molecular characterization of sponges by sequencing of standard genetic markers – known as DNA barcoding – is being used increasingly as a means to facilitate identification and to complement the description of new species [37]. Almost invariably, analyses involve the use of the subunit I of the cytochrome C oxidase gene (CO1) [37]–[39]. The genetic variation (p-distance) found in our host species’ CO1 gene, i.e. no intraspecific variation and a 4.7% genetic distance between I. variabilis and S. spinosulus, are within the range of values observed for other Irciniids using the same marker. In a barcoding study of Indo-Pacific Irciniids, Pöppe et al. [40] observed no intraspecific variation within any of the analysed species, low interspecific variation between congeners (0.2–2.7% in Ircinia spp. and 0.2–1.7% in Psammocinia spp.), and higher differentiation levels between members of the two genera (p-distances of 3.1–5.8%) [40]. In a second study comparing the bacterial symbionts in three species of Ircinia in the Mediterranean Sea, Erwin et al. [18] found no intraspecific variation within any of the studied species (nor between I. variabilis/fasciculata) and a p-distance of 0.6–1.8% between the different species. Not unexpectedly we found some genetic distance (p-distance 0–0.6%) between the sequences of our Atlantic specimens and those available on GenBank from Mediterranean specimens. This may indicate some level of genetic isolation and differentiation between conspecific populations occurring in these areas as previously observed in other sponge taxa (e.g. Xavier, et al. [41]). Overall, host phylogenetic inference can be a suitable and complementary tool in sponge microbiology studies - as shown in early [36], [42] and recent [18], [43] reports on host-symbiont co-evolutionary relationships. Its use seems especially well suited to the study of sponge hosts displaying smooth gradients of phylogenetic relatedness or unresolved taxonomies such as the members of the Irciniidae family and its relevance in such studies is likely to rise with the analysis of multiple phylogenetic markers in concatenation.
In the present survey, the abundance of culturable bacteria associated with S. spinosulus and I. variabilis was similar. It is well-known that many aspects affect bacterial cultivation and the use of standard culture media has so far allowed the assessment of only a minor fraction (e.g. from 0.1 to 1%) of bacteria associated with marine sponges [44]–[46]. This might sharply compromise the comparative assessment of bacterial abundance in sponges when solely using this technique. To circumvent the limitations inherent from cultivation, epifluorescence microscopy was applied to estimate abundance by enabling the count of all detectable nucleic-acid containing cells present in the samples. Based on the cell counts retrieved with this method, about 3 orders of magnitude higher than the registered CFU counts, S. spinosulus and I. variabilis can be regarded as HMA sponges, supporting previous observations obtained for tropical Irciniidae species such as I. felix and I. strobilina [12], [47], [48].
Bacteria, Actinobacteria and Alphaproteobacteria PCR-DGGE fingerprinting revealed a clear difference in bacterial diversity and community composition between sponge and seawater samples. This expected trend has been reported in several previous sponge microbiology surveys [1], [49], [50]. In agreement with our results, the bacterial PCR-DGGE profiles from I. felix collected at two sites in Key Largo, Florida, revealed distinct band patterns in comparison with seawater samples [12], whereas bacterial PCR-DGGE profiles of wild and captivated I. strobilina specimens were likewise distinct when compared with surrounding seawater and water used in sponge aquaculture, respectively [15]. In contrast with species of the genus Ircinia, knowledge of bacterial abundance and diversity in Sarcotragus specimens is virtually nonexistent. Here we showed S. spinosulus-specific profiles that strongly differed from those found in I. variabilis, with several species-specific PCR-DGGE bands detected and further identified (see below). S. spinosulus exhibited greater bacterial community diversity and richness, and homogeneity across individual specimens than the latter. Further, evidence for greater prokaryotic abundance in S. spinosulus was found. In a recent survey, Erwin et al. [18] could detect bacterial OTUs exclusive to the species I. oros, I. fasciculata or I. variabilis in the Mediterranean Sea. Taken together, these studies hint at a fundamental role of the host in shaping the structure and promoting diversity of symbiont communities within closely related sponge hosts. Interestingly, functional equivalence and evolutionary convergence of symbiont communities have been suggested as an evolutionary model applicable to the complex sponge microbiota, based on the share of core microbial functions between six phylogenetically distant sponge species with different symbiont community structures [51]. In this context, it is tempting to speculate that the less studied Sarcotragus also establishes close interactions with selected bacterial communities, which regardless their degree of distinctiveness might have intrinsic functions like those observed for Ircinia spp. [11], [15], [16]. Future studies addressing microbial functioning in sympatric and phylogenetically close hosts will certainly shed further light on our current understanding of symbiont evolution within sponges.
We successfully identified several sponge-specific bacterial populations by PCR-DGGE. Four dominant symbionts - two from S. spinosulus, one from I. variabilis and one found in both sponge species - were affiliated with an uncultured actinobacterial lineage within the order Acidimicrobiales [52]. Three of these (“bands” 3, 6 and 7 in Figs. 3 and 4) belonged to a cluster of sponge-specific sequences collected worldwide and called SC22 by Simister et al. [34], whereas the fourth grouped into a different and diverse cluster dominated by sponge-derived bacterial sequences (Fig. 4). Within cluster SC22, sequences were obtained from adult of Svenzea zeai and Smenospongia aurea along with their reproductive material, which suggests that vertical transmission of this particular phylotype is likely to occur [53], [54]. The same observation was made for the cluster formed by the fourth symbiont in the Acidimicrobiales group (“band 1” in Fig. 4) and related sequences, from which two sequences from adult S. zeai and one from its embryo were found [53]. These results indicate an intimate pattern of relationship between sponge-associated Acidimicrobiales and their hosts. The order Acidimicrobiales contains mesophilic and moderate thermophilic species and all members are obligatory acidophilic found in iron-, sulphur- or mineral-sulfide rich environments. Species within this order are capable of ferrous iron and sulphur oxidation and ferric iron reduction [55]–[58]. However, the physiological properties exhibited by cultivated Acidimicrobiales might not necessarily match those of marine sponge symbionts, as these usually share lower relatedness to cultured species at the 16S rRNA gene level, and therefore further research is needed to unveil the ecology and functioning of these symbionts in marine sponges.
A prevailing symbiont found exclusively in S. spinosulus was affiliated with uncultured Gammaproteobacteria within the order Chromatiales [59]. These sequences belonged to a cluster of sponge-specific sequences acquired worldwide (Fig. 5). Among them, adult sequences from Ircinia felix, Smenospongia aurea and Svenzea zeai were observed along with sequences from reproductive material of I. felix, S. aurea and Ectyoplasia ferox [11], [53], [54]. The order Chromatiales encompasses members of the purple sulphur bacteria that are capable of performing anoxygenic photosynthesis using hydrogen sulphide as electron donor [59]. Furthermore, many Chromatiales species have been shown to perform fixation of molecular nitrogen [59], [60]. These functions might be highly valuable for sponge survival, and the consistency with which members of this group are found in marine sponges at a global scale indeed suggests that Chromatiales species play an important role in their association with such hosts.
Another phylotype solely recovered from S. spinosulus was affiliated with an uncultured, sponge-specific lineage in the Chloroflexi phylum, named SC46 by Simister et al. [34] (Fig. 6). The Chloroflexi is regarded as one of the most abundant and diverse bacteria phyla associated with a wide variety of marine sponges, with many sponge-specific clusters identified [6], [34], [61]. So far, only one Chloroflexi species was isolated from the sponge Geodia spp., which also clustered with sequences exclusively obtained from marine sponges [62]. In shallow waters, members of Chloroflexi are able to fix atmospheric carbon through photosynthesis, and thus these bacteria could provide carbonaceous compounds to the sponge host [62]. Recently, a Chloroflexi bacterium was pointed as the likely producer of a novel non-ribosomal peptide synthase [63]. Thus, Chloroflexi strains might play important roles in sponge nutrition and defence.
Using a taxon-specific fingerprinting approach to the Alphaproteobacteria, a dominant symbiont exclusive to S. spinosuls was uncovered (“band 8” in Fig. 3) and found to be closely related to an uncultured alphaproteobacterium within the family Rhodobacterales [64]. Sequences representing this symbiont formed a concise cluster with sequences retrieved from marine sponges in several geographical backgrounds in addition to cultured representatives obtained from different environments such as microbial mats, seawater, soil from saltpan, water and marine aquaculture pond [65], [66] (Fig. 7). This clade contained sequences obtained from adults of Xestospongia muta and Svenzea zeai along with their reproductive material [53], [54]. This symbiont is closely related to Rhodovulum species, in which many type strains have been mostly retrieved from marine habitats. This genus contains species that undertake diverse metabolic pathways such as photoautotrophic, photoheterotrophic and chemotrophic and occur mainly in marine and hypersaline environments under oxic, microoxic and anoxic conditions [67]. The metabolic versatility of Rhodovulum species indicate that they are able to use the waste generated by sponges. For instance, ammonia, which is a toxic metabolic waste product that could accumulate within the sponge body, especially during low pumping activity, might be used as nitrogen source for Rhodovulum species [1], [67], [68]. In addition, some strains of Rhodovulum could be involved in nitrogen and sulphur cycling, once they are capable to use dinitrogen, sulphur, sulphite, sulphate and thiosulfate [67]. Vertical transmission has also been documented for members of this genus in marine sponges, [54] suggesting Rhodovulum as a likely, relevant constituent of the sponge-associated microbiome.
The present study provides first insights into the bacterial abundance and diversity in Atlantic S. spinosulus and I. variabilis. In spite of their sympatric occurrence, the inspected species hosted bacterial communities that differ from each other and from those found in seawater. Interestingly, all bands excised from PCR-DGGE profiles that were exclusive to sponge samples affiliated with previously identified sponge-specific sequence clusters [34] or with potentially novel sponge-specific clusters found in the present survey. Thus, the approach used here enabled not only straightforward assessment of overall trends in bacterial community structures, but also direct identification of symbionts of putative relevance in association with their hosts, given their dominance and consistent patterns of occurrence in the analysed specimens, and their presumed sponge-specific life histories as inferred by 16S rRNA gene phylogenies. Notably, bacterial phylotypes regarded as “S. spinosulus-specific” or “I. variabilis-specific” in this study shared high degrees of resemblance with sponge-derived sequences from other biogeographical settings and/or more distantly related sponge hosts. This picture, in which bacterial signatures not shared by co-occurring and taxonomically close sponge species are found in disparate sponge hosts and localities, most likely derives from factors of the host and of the environment – including vertical transmission vs. environmental acquisition of symbionts, specific habitat preferences and life stages of the host - that cooperatively shape the structure of the sponge-associated microbiome [1], [18]. As a result, complex communities of specific composition at the host species or even specimen level [9], [28] with concomitant sharing, across sponge species, of generalist symbionts displaying broad host range and/or widespread occurrence [43], [69] have often been reported for marine sponges. Here, the distinct communities observed in S. spinosulus and I. variabilis within the same habitat, along with the detection of symbionts showing broad host and geographical ranges as inferred by 16S rRNA gene phylogenies, hints at a pivotal role for the host in shaping the structure of its own microbiota while revealing versatile and widespread bacterial phylotypes with apparently intimate sponge-associated life histories. The high abundance and species-specific character of these assemblages suggest in-faunal microbial communities as overriding drivers of functioning and of genetic and metabolic diversities in coastal ecosystems.
Materials and Methods
Sponge and seawater sampling
Four specimens of Sarcotragus spinosulus and Ircinia variabilis (Demospongiae, Dictyoceratida, Irciniidae) were collected by scuba diving at depths around 15 m at Galé Alta, Armação de Pêra (37° 04′ 09.6′′N and 8° 19′ 52.1′′W) in the coast of the Algarve, Portugal, in June 2010. Measurements of temperature, oxygen and salinity during the sampling procedure were 14.6°C, 5.95 mg L−1 and 35.11 part per million (ppm), respectively. In situ pictures of the specimens were taken to aid laboratory identification (Fig. S1). The individual samples were placed, in situ, separately in plastic bags (type ziploc®) containing natural seawater, transferred into cooling boxes, brought to the laboratory within few hours and processed upon arrival. Four samples of seawater (1L each) from the vicinity of the sponges (i.e. 1 m above the specimens) were also collected as above. Prior to sample processing, the sponge specimens were rinsed with sterile Artificial Seawater (ASW) [70] to remove loosely associated organisms. Voucher samples were preserved in 90% ethanol for taxonomic identification and deposited in the Biology Department´s zoological collection of the University of the Azores (DBUA.Por). Because sampling did not involve endangered or protected species and did not occur within privately owned or protected areas, no specific permits were required for the described field studies.
Sponge identification
Specimens were identified from the analysis of general external morphological characters and internal skeletal features, i.e. thickness, degree of fasciculation and presence of foreign debris within the spongin fibres and structure of the collagenous filaments. Genera within the family Irciniidae are distinguished by the presence of a cortical armour of sand (exclusive to Psammocinia), and presence (in Ircinia) versus absence (in Sarcotragus) of foreign debris within the primary fibres [35].
Phylogenetic inference of sponge specimens (commonly referred to as “sponge DNA barcoding”) was used to aid species identification by molecular means. PCR amplifications were carried out on sponge total community DNA (see below) targeting the subunit I of the cytochrome oxidase gene with the primers dgLCO1490 and dgHCO2189 [71]. This fragment (c. 640 bp) encompasses the standard “barcoding” partition [72]. The reaction mixture (25 µL) contained 1.5 µL of template DNA (∼20 ng), 1X reaction buffer (Bioline, London, UK), 0.16 mM deoxynucleoside triphosphates (dNTPs), 4.0 mM MgCl2, 0.64 mg mL−1 of bovine serum albumin (BSA), 0.24 µM of primers and 0.625U of BioTaq™ DNA polymerase (Bioline, London, UK). After initial denaturation at 95°C for 3 min., 36 cycles of 45 sec at 94°C, 60 sec at 51°C and 90 sec at 72°C were carried out. A final extension of 10 min at 72°C was used to finish the reaction. All PCR amplifications were carried out in a MyCycle thermal cycler (Bio-Rad, Hercules, CA, USA). Amplicons were checked after electrophoresis on 1% agarose gel under UV light. PCR products with right size were cleaned with Sephadex G50 (GE Healthcare Bio-Science AB, Uppsala, Sweden) columns, quantified with Image Lab™ Software (Bio-Rad, Hercules, CA, USA), and subjected to sequencing with the chain termination method in an Applied Biosystems 3130 genetic analyser using the forward primer. Closest relatives were searched using the megablast and blastn algorithms of the National Center for Biotechnology Information (NCBI) [73]. Closely related sequences from the NCBI and the Sponge Barcoding Project (www.spongebarcoding.org) databases were used to retrieve representative CO1 sequences for phylogenetic inference (see below).
Plate counting of heterotrophic bacteria
Per sponge specimen, 2.5 g of fresh internal body was cut and transferred to a 50 mL screw cap polypropylene tube containing 25 mL of Calcium/Magnesium Free Artificial Seawater (CMFASW) [74]. The sponge samples were ground with sterile mortar and pestle. The resulting suspensions were collected and allowed to decant for 5 min. Serial 10-fold dilutions were then prepared with sterile ASW and plated in triplicate onto Marine broth (Carl Roth GmbH+Co, Germany) plus 1.5% agar. The plates were incubated at room temperature (∼25°C) and Colony Forming Unit (CFU) counting was performed after 3, 5 and 7 days of incubation. Log-transformed CFU values fitted the normal distribution and were compared by One Way Analysis of Variance (ANOVA) using PASW Statistics 18 (SPSS Inc., Chicago, USA).
Epifluorescence microscopy
A cultivation-independent analysis of prokaryotic abundance based on epifluorescence microscopy was performed in this study. For the sponge samples, the suspensions prepared in the abovementioned procedure were first centrifuged at 500 g for 2 min to remove sponge cells and debris. Aliquots (100 µL) of the resulting supernatants were individually fixed in 2.5% glutaraldehyde and the volume was completed to 10 mL with sterile ASW. Seawater samples (9.2 mL) were also fixed in 2.5% glutaraldehyde. From the fixed material, 100 µL and 10 mL from sponge and seawater samples, respectively, were filtered through 0.2-µm-pore-size isopore™ black membrane filters (Millipore, Bellerica, MA, USA). The filter was stained with the DNA-binding fluorochrome acridine orange, mounted on glass slides and analysed with an inverted research system microscope IX81 (Olympus Europa GmbH, Hamburg, Germany) where 25 photos per specimen were taken at random. Cells with a well-defined edge, usually ranging from 0.2 to 1 µm in diameter when coccoid, or reaching up to 5 µm in length when rod-shaped, were counted and served as proxy for prokaryotic cell abundance in the samples. Larger objects (>5 µm) that could eventually account for eukaryotic organisms were not considered. Total prokaryotic numbers were log-transformed and analysed by One Way ANOVA using PASW Statistics 18 (SPSS Inc., Chicago, USA).
Total community DNA extraction
Genomic DNA of about 0.25 g of internal sponge body was extracted using UltraClean® Soil DNA isolation kit (Mo Bio, Carlsbad, CA, USA) according to the manufacturer’s protocol. Based on preliminary PCR-DGGE assessments, this method led to a more reproducible depiction of bacterial community structures in the sponges when compared with a method that employs a cell-separation treatment prior to DNA extraction (Hardoim et al., unpublished results), and was therefore chosen for the purpose of this study. Seawater samples (500 mL) were filtered through 0.2-µm-pore-size nitrocellulose filters (Millipore, Billerica, MA, USA) using a vacuum pump. The filters were cut into small pieces and directly used for DNA extraction as explained above.
Bacterial PCR for DGGE analysis
A nested PCR-denaturing gradient gel electrophoresis (PCR-DGGE) approach was chosen - based on its higher detection sensitivity and reproducibility when compared with a one-step amplification protocol in preliminary assays (data not shown) - to assess the total bacterial communities in all samples. Nearly full-length 16S rRNA gene fragments were amplified with the primer pair F27 and R1492 [75]. The reaction mixture (25 µL) was prepared with 1 µL of template DNA (∼20 ng), 1X Stoffel buffer (Applied Biosystems, Foster, CA), 0.2 mM dNTPs, 3.75 mM MgCl2, 0.1 mg mL−1 of BSA, 2% (vol/vol) dimethyl sulfoxide (DMSO), 0.2 µM of each primer, and 1.25U of Taq DNA polymerase (Applied Biosystems, Foster, CA). After initial denaturation at 94°C for 5 min, 30 cycles of 1 min at 94°C, 1 min at 56°C and 2 min at 72°C were performed, followed by a final extension for 10 min at 72°C. The amplicons (1.5 µL) were used as template in a subsequent PCR for DGGE analysis (20 cycles) using the primer pair F984-GC and R1378 [76]. The PCR mixture and thermal cycling followed the protocol by Costa et al. [77], with half the quantity of Taq DNA polymerase (1.25 U) per reaction.
PCR of Specific Bacterial Groups for DGGE Analyses
Actinobacteria 16S rRNA gene fragments
The first amplification of the nested PCR was carried out with the primers F243 [76] and R1494 [75] to generate Actinobacteria-specific amplicons. The reaction mixture and PCR conditions were carried out as described by Hardoim et al. [6], except for the concentration of Taq DNA polymerase (1.25U), number of cycles (25 cycles) and extension period (1 min) in the present study. The amplicons (2 µL) were used in a second PCR for DGGE analysis using the primers F984-GC and R1378 [76] as described previously for total bacteria, except for the number of cycles (30 cycles).
Alphaproteobacteria 16S rRNA gene fragments
The first reaction mixture of the nested PCR was prepared as described by Gomes et al. [78], except that in the present study the primer concentration and Taq DNA polymerase were 0.2 µM and 1.25 U, respectively, and that BSA was not used in the group-specific PCR. After initial denaturation at 94°C for 7 min, 30 cycles of 1 min at 94°C, 1 min at 56°C and 1 min at 72°C were carried out. The reaction was finished with an extension of 10 min at 72°C. Amplicons from the first reaction (2 µL) were used in a subsequent PCR for DGGE analysis as described previously, except for the number of cycles (25 cycles).
PCR-DGGE profiling
DGGE assays were carried out in a PhorU-2 gradient system (Ingeny International, Goes, The Netherlands). The 16S rRNA gene amplicons generated as explained above were applied in even concentrations onto polyacrylamide gels containing a 46.5 to 65% gradient of denaturants (100% denaturants defined as 7 M urea and 40% formamide) and a 6.2 to 9% gradient of acrylamide. Mixtures of PCR products of ten bacterial strains isolated from Sarcotragus sp. and Ircinia sp. (Staphylococcus sp.; Ruegeria sp.; Pseudomonas sp.; Leisingera sp.; Corynebacterium sp.; Micrococcus sp.; Streptomyces sp. and Pontibacter sp.) were loaded at the edge of the gels as markers. Electrophoresis was performed in a 1X Tris-acetate-EDTA buffer (pH 7.8) at 58°C and 140V for 16 h. The gels were silver stained [76] and air dried, after which digital images were obtained by scanning.
Analysis of PCR-DGGE profiles
The software GelCompar II 5.1 (Applied Maths, Kortrijk, Belgium) was used to analyse the PCR-DGGE profiles as recommended by Rademaker and de Bruijn, [79]. Briefly, pairwise Pearson correlation coefficients (r) were calculated as a measurement of the similarity between the community profiles. Cluster analysis was carried out with the unweighted pair group method with mathematical averages (UPGMA) using the similarity matrix generated with the calculated Pearson coefficients. In addition to cluster analysis, constrained (i.e. canonical) ordination was performed with the Canoco for Windows 4.5 software package (Microcomputer Power. Ithaca, NY) using a “sample×species” datasheet as input, in which the “species” data represent the presence and relative abundance of PCR-DGGE bands in each fingerprint, as described in detail by Costa et al. [80]. This was used to infer whether sponge species and seawater significantly contributed to the observed variability in the PCR-DGGE profiles (see [6], [80]). The Shannon measure of diversity (H’), determined as H’ = –∑ pi.logpi where pi represents the relative abundance of the ith category (i.e. PCR-DGGE band) within the sample (i.e. PCR-DGGE fingerprint) was applied to estimate the diversity of each PCR-DGGE fingerprint generated in this study. The evenness (J’ = H’/Hmax) of PCR-DGGE fingerprints was calculated based on the diversity indices obtained. Measures of richness (i.e. number of PCR-DGGE bands), diversity and evenness of PCR-DGGE fingerprints were compared by One Way ANOVA using PASW Statistics 18 (SPSS Inc., Chicago, USA).
Identification of dominant bands in PCR-DGGE profiles
Sponge-associated and seawater exclusive bands were visually determined based on their occurrence across replicates (i.e. only bands detected in at least 3 of 4 replicates were sequenced). Further, their discriminating power was assessed, and only bands displaying high variance in relative abundance as a response to the sample classes “I. variabilis”, “S. spinosulus” and “Seawater” were selected. Discriminating bands were revealed by the species fit range function in the Canoco for windows 4.5 software, where only those bands displaying 50% fit range or more were considered for sequencing purposes. Discriminating bands were excised from DG-gels and re-amplified for PCR-DGGE analysis using the method of Costa et al. [80]. The resulting amplicons were loaded onto DGGE with the original community DNA samples to verify their electrophoretic mobility. Excised bands that displayed the right melting behaviour when compared with the original band in the community profiles were used as templates in another PCR amplification, in which the forward primer F984 used had no GC clamp. PCR-DGGE reaction mixtures and thermal cycles were carried out as described above. The amplicons were then purified in Sephadex G50 columns, quantified with Image Lab™ Software, and subjected to sequencing as above mentioned. For some excised bands, no pure amplicon was recovered and thus a cloning procedure was undertaken using the pGEM-T Vector System II Kit (Promega, Madison, WI) as described elsewhere [6], [77], [80]. Clones that showed the same electrophoretic mobility when compared to their original band were selected for sequencing as explained above. All sequences retrieved in this study were submitted to the EMBL Nucleotide Sequence Database under accession numbers HE797930 to HE797937 for sponge CO1 sequences and HE797938 to HE797958 for PCR-DGGE bands representing bacterial 16S rRNA genes.
Phylogenetic analyses
Sequences generated from sponge CO1 amplification and excised bacterial bands were quality-inspected and edited with the Sequence Scanner software V.1 (Applied Biosystems). For bacterial DGGE bands, taxonomic assignment of sequences was performed with the seqmatch and classifier tools of the Ribosomal Database Project II (http://rdp.cme.msu.edu) at 80% confidence threshold. Closest phylogenetic relatives were searched with the blast-n algorithm of NCBI. PCR-DGGE band sequences and their closest phylogenetic relatives were aligned using the SINA web aligner [81]. Aligned sequences were then imported into a modified SILVA 16S rRNA gene database version 102, which included all sponge-derived 16S rRNA gene sequences available in early 2010 [34], using the parsimony tool as implemented in the ARB software [82]. The sponge database generated by Simister et al. [34] contained phylogenetic inferences performed with long sequences (≥1200bp) using the program RAxML for all sponge-associated bacterial phyla, from which sponge-specific clusters were assigned [34] according to the criteria described by Hentschel et al. [69]. Alignments were manually checked and corrected when necessary using the ARB alignment window. The sequences generated in this study were added to maximum likelihood trees inferred by Simister et al. [34] through the parsimony interactive tool available in ARB using 50% conservation filters for each of the corresponding bacterial phyla, and their affiliation with sponge-specific phylogenetic clusters was then ascertained. From the resulting trees, relevant sequences were selected for further phylogenetic analyses (see below). The CO1 gene sequences from each investigated specimen were aligned against selected sponge barcoding sequences using Clustal X in the MEGA5 software [83]. Phylogenetic inferences of bacteria and sponge sequences were performed as described by Hardoim et al. [84]. Briefly, an appropriate evolutionary model for all phylogenetic trees was determined using ModelGenerator version 85 [85] and found to be the general-time reversible model (GTR, [86] with a discrete gamma-distribution of among-site rate variation (Γ4) and a proportion of invariant sites (I), except for CO1 inference, in which invariant sites did not fit. Maximum likelihood and Bayesian MCMC analyses were conducted using RAxML (vers. 7.0.4-MPI) and MrBayes (vers. 3.2.1), respectively [87]–[89].
Supporting Information
Sponge species. In situ pictures of S. spinosulus (A) and I. variabilis (B).
(TIF)
Cluster analysis. Cluster analysis of PCR-DGGE fingerprints obtained for Bacteria (A), Actinobacteria (B) and Alphaproteobacteria (C). S. spinosulus: Alg10/08, Alg10/09, Alg10/10 and Alg10/11; I. variabilis: Alg10/12, Alg10/13, Alg10/14 and Alg10/15 and Seawater: SW07, SW22, SW23 and SW24.
(TIF)
PCR-DGGE band richness, diversity and evenness.
(DOCX)
Acknowledgments
We acknowledge Msc. Sandra Mesquita, Dr Paulo J. Gavaia and João C. Silva from the Centre of Marine Sciences (CCMar-UAlg) for their help in epifluorescence microscopy sample preparation and analysis. We thank Dr. Christin Zachow from the Institute of Environmental Biotechnology, Graz University of Technology, Austria, for her help with Gelcompar analysis. We are grateful to Dr. Michael Taylor for kindly providing the ARB database containing sponge-derived 16S rRNA gene sequences [34] used in this study to diagnose sponge-specific clusters.
Funding Statement
This work was financed by the Portuguese Foundation for Science and Technology (FCT - http://www.fct.pt) through the research project PTDC/MAR/101431/2008. CCPH has a PhD fellowship granted by FCT (Grant No. SFRH/BD/60873/2009). JRX's research is funded by a FCT postdoctoral fellowship (grant no. SFRH/BPD/62946/2009). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Taylor MW, Radax R, Steger D, Wagner M (2007) Sponge-associated microorganisms: Evolution, ecology, and biotechnological potential. Microbiol Mol Biol R 71: 295–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Henstchel U, Piel J, Degnan SM, Taylor MW (2012) Genomic insights into the marine sponge microbiome. Nat Rev Microbiol 10: 641–654. [DOI] [PubMed] [Google Scholar]
- 3. Vacelet J, Donadey C (1977) Electron-microscope study of association between some sponges and bacteria. J Exp Mar Biol Ecol 30: 301–314. [Google Scholar]
- 4. Friedrich AB, Merkert H, Fendert T, Hacker J, Proksch P, et al. (1999) Microbial diversity in the marine sponge Aplysina cavernicola (formerly Verongia cavernicola) analyzed by fluorescence in situ hybridization (FISH). Mar Biol 134: 461–470. [Google Scholar]
- 5. Hentschel U, Usher KM, Taylor MW (2006) Marine sponges as microbial fermenters. FEMS Microbiol Ecol 55: 167–177. [DOI] [PubMed] [Google Scholar]
- 6. Hardoim CCP, Costa R, Araujo FV, Hajdu E, Peixoto R, et al. (2009) Diversity of bacteria in the marine sponge Aplysina fulva in Brazilian coastal waters. Appl Environ Microb 75: 3331–3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Thomas TRA, Kavlekar DP, LokaBharathi PA (2010) Marine drugs from sponge-microbe association - A Review. Mar Drugs 8: 1417–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Webster NS, Taylor MW, Behnam F, Lücker S, Rattei T, et al. (2010) Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts. Environ Microbiol 12: 2070–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee OO, Wang Y, Yang JK, Lafi FF, Al-Suwailem A, et al. (2011) Pyrosequencing reveals highly diverse and species-specific microbial communities in sponges from the Red Sea. ISME J 5: 650–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Usher K, Kuo J, Fromont J, Toze S, Sutton D (2006) Comparative morphology of five species of symbiotic and non-symbiotic coccoid Cyanobacteria . Eur J Phycol 41: 179–188. [Google Scholar]
- 11. Schmitt S, Weisz J, Lindquist N, Hentschel U (2007) Vertical transmission of a phylogenetically complex microbial consortium in the viviparous sponge Ircinia felix . Appl Environ Microb 73: 2067–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Weisz J, Hentschel U, Lindquist N, Martens C (2007) Linking abundance and diversity of sponge-associated microbial communities to metabolic differences in host sponges. Mar Biol 152: 475–483. [Google Scholar]
- 13. Mohamed NM, Colman AS, Tal Y, Hill RT (2008) Diversity and expression of nitrogen fixation genes in bacterial symbionts of marine sponges. Environ Microbiol 10: 2910–2921. [DOI] [PubMed] [Google Scholar]
- 14. Mohamed N, Cicirelli E, Kan J, Chen F, Fuqua C, et al. (2008) Diversity and quorum-sensing signal production of Proteobacteria associated with marine sponges. Environ Microbiol 10: 75–86. [DOI] [PubMed] [Google Scholar]
- 15. Mohamed N, Rao V, Hamann M, Kelly M, Hill R (2008) Monitoring bacterial diversity of the marine sponge Ircinia strobilina upon transfer into aquaculture. Appl Environ Microb 74: 4133–4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mohamed NM, Saito K, Tal Y, Hill RT (2010) Diversity of aerobic and anaerobic ammonia-oxidizing bacteria in marine sponges. ISME J 4: 38–48. [DOI] [PubMed] [Google Scholar]
- 17. Yang JK, Sun J, Lee OO, Wong YH, Qian PY (2011) Phylogenetic diversity and community structure of sponge-associated bacteria from mangroves of the Caribbean Sea. Aquat Microb Ecol 62: 231–240. [Google Scholar]
- 18. Erwin PM, López-Legentil S, Gonzalez-Pech R, Turon X (2012) A specific mix of generalists: bacterial symbionts in Mediterranean Ircinia spp. FEMS Microbiol Ecol 79: 619–637. [DOI] [PubMed] [Google Scholar]
- 19. Cichewicz R, Valeriote F, Crews P (2004) Psymberin, a potent sponge-derived cytotoxin from Psammocinia distantly related to the pederin family. Org Lett 6: 1951–1954. [DOI] [PubMed] [Google Scholar]
- 20. Emura C, Higuchi R, Miyamoto T (2006) Irciniasulfonic acid B, a novel taurine conjugated fatty acid derivative from a Japanese marine sponge, Ircinia sp. Tetrahedron 62: 5682–5685. [Google Scholar]
- 21. Liu Y, Zhang S, Abreu P (2006) Heterocyclic terpenes: linear furano- and pyrroloterpenoids. Nat Prod Rep 23: 630–651. [DOI] [PubMed] [Google Scholar]
- 22. Xu S, Liao X, Du B, Zhou X, Huang Q, et al. (2008) A series of new 5,6-epoxysterols from a Chinese sponge Ircinia aruensis . Steroids 73: 568–573. [DOI] [PubMed] [Google Scholar]
- 23. Wätjen W, Putz A, Chovolou Y, Kampkötter A, Totzke F, et al. (2009) Hexa-, hepta- and nonaprenylhydroquinones isolated from marine sponges Sarcotragus muscarum and Ircinia fasciculata inhibit NF-kappa B signalling in H4IIE cells. J Pharm Pharmacol 61: 919–924. [DOI] [PubMed] [Google Scholar]
- 24. Orhan I, Sener B, Kaiser M, Brun R, Tasdemir D (2010) Inhibitory activity of marine sponge-derived natural products against parasitic Protozoa. Mar Drugs 8: 47–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. De Rosa S, Mitova M, Tommonaro G (2003) Marine bacteria associated with sponge as source of cyclic peptides. Biomol Eng 20: 311–316. [DOI] [PubMed] [Google Scholar]
- 26. Mitova M, Tommonaro G, De Rosa S (2003) A novel cyclopeptide from a bacterium associated with the marine sponge Ircinia muscarum . Z Naturforschung C 58c: 740–745. [DOI] [PubMed] [Google Scholar]
- 27. Fisch K, Gurgui C, Heycke N, van der Sar S, Anderson S, et al. (2009) Polyketide assembly lines of uncultivated sponge symbionts from structure-based gene targeting. Nat Chem Biol 5: 494–501. [DOI] [PubMed] [Google Scholar]
- 28. Schmitt S, Tsai P, Bell J, Fromont J, Ilan M, et al. (2011) Assessing the complex sponge microbiota: core, variable and species-specific bacterial communities in marine sponges. ISME J 6: 564–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kvennefors ECE, Sampayo E, Ridgway T, Barnes AC, Hoegh-Guldberg O (2010) Bacterial communities of two ubiquitous Great Barrier Reef corals reveals both site-and species-specificity of common bacterial associates. PLoS ONE 5: e10401 doi:10.1371/journal.pone.0010401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fierer N, Lauber CL, Zhou N, McDonald D, Costello EK, et al. (2010) Forensic identification using skin bacterial communities. P Natl Acad Sci USA 107: 6477–6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, et al. (2011) Enterotypes of the human gut microbiome. Nature 473: 174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bull AT, Stach JEM (2007) Marine Actinobacteria: new opportunities for natural product search and discovery. Trends Microbiol 15: 491–499. [DOI] [PubMed] [Google Scholar]
- 33. Webster NS, Taylor MW (2012) Marine sponges and their microbial symbionts: love and other relationships. Environ Microbiol 14: 335–346. [DOI] [PubMed] [Google Scholar]
- 34. Simister RL, Deines P, Botté ES, Webster NS, Taylor MW (2012) Sponge-specific clusters revisited: a comprehensive phylogeny of sponge-associated microorganisms. Environ Microbiol 14: 514–524. [DOI] [PubMed] [Google Scholar]
- 35.Cook SC, Bergquist PR (2002) Family Irciniidae Gray, 1867, In: Hooper JNA, van Soest RWM, editors. System Porifera: a guide to the classification of sponges. Kluwer Academic/Plenum Publishers, New York, NY. 1022–1027.
- 36. Erpenbeck D, Breeuwer J, van der Velde H, van Soest R (2002) Unravelling host and symbiont phylogenies of halichondrid sponges (Demospongiae, Porifera) using a mitochondrial marker. Mar Biol 141: 377–386. [Google Scholar]
- 37. Wörheide G, Erpenbeck D (2007) DNA taxonomy of sponges - progress and perspectives. J Mar Biol Assoc UK 87: 1629–1633. [Google Scholar]
- 38. Erpenbeck D, Duran S, Rutzler K, Paul V, Hooper JNA, et al. (2007) Towards a DNA taxonomy of Caribbean demosponges: a gene tree reconstructed from partial mitochondrial CO1 gene sequences supports previous rDNA phylogenies and provides a new perspective on the systematics of Demospongiae. J Mar Biol Assoc UK 87: 1563–1570. [Google Scholar]
- 39.Cardenas P, Menegola C, Rapp HT, Diaz MC (2009) Morphological description and DNA barcodes of shallow-water Tetractinellida (Porifera: Demospongiae) from Bocas del Toro, Panamá, with description of a new species. Zootaxa: 1–39.
- 40. Pöppe J, Sutcliffe P, Hooper JNA, Wörheide G, Erpenbeck D (2010) CO I barcoding reveals new clades and radiation patterns of Indo-Pacific sponges of the Family Irciniidae (Demospongiae: Dictyoceratida). PLoS ONE 5: e9950 doi:10.1371/journal.pone.0009950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xavier JR, Van Soest RWM, Breeuwer JAJ, Martins AMF, Menken SBJ (2010) Phylogeography, genetic diversity and structure of the poecilosclerid sponge Phorbas fictitius at oceanic islands. Contrib Zool 79: 119–129. [Google Scholar]
- 42. Thacker R, Starnes S (2003) Host specificity of the symbiotic cyanobacterium Oscillatoria spongeliae in marine sponges, Dysidea spp. Mar Biol 142: 643–648. [Google Scholar]
- 43. Montalvo NF, Hill RT (2011) Sponge-associated bacteria are strictly maintained in two closely related but geographically distant sponge hosts. Appl Environ Microb 77: 7207–7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Santavy DL, Willenz P, Colwell RR (1990) Phenotypic study of bacteria associated with the caribbean sclerosponge, Ceratoporella nicholsoni . Appl Environ Microb 56: 1750–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Friedrich AB, Fischer I, Proksch P, Hacker J, Hentschel U (2001) Temporal variation of the microbial community associated with the mediterranean sponge Aplysina aerophoba . FEMS Microbiol Ecol 38: 105–113. [Google Scholar]
- 46. Webster NS, Hill RT (2001) The culturable microbial community of the Great Barrier Reef sponge Rhopaloeides odorabile is dominated by an alpha-Proteobacterium. Mar Biol 138: 843–851. [Google Scholar]
- 47. Vicente VP (1990) Response of sponges with autotrophic endosymbionts during the coral-bleaching episode in Puerto Rico. Coral Reefs 8: 199–202. [Google Scholar]
- 48. Weisz JB, Lindquist N, Martens CS (2008) Do associated microbial abundances impact marine demosponge pumping rates and tissue densities? Oecologia 155: 367–376. [DOI] [PubMed] [Google Scholar]
- 49. Taylor MW, Schupp PJ, Dahllof I, Kjelleberg S, Steinberg PD (2004) Host specificity in marine sponge-associated bacteria, and potential implications for marine microbial diversity. Environ Microbiol 6: 121–130. [DOI] [PubMed] [Google Scholar]
- 50. Taylor MW, Schupp PJ, de Nys R, Kjelleberg S, Steinberg PD (2005) Biogeography of bacteria associated with the marine sponge Cymbastela concentrica . Environ Microbiol 7: 419–433. [DOI] [PubMed] [Google Scholar]
- 51. Fan L, Reynolds D, Liu M, Stark M, Kjelleberg S, et al. (2012) Functional equivalence and evolutionary convergence in complex communities of microbial sponge symbionts. P Natl Acad Sci USA 109: E1878–E1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stackebrandt E, Rainey FA, Ward-Rainey NL (1997) Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int J Syst Bacteriol 47: 479–491. [Google Scholar]
- 53. Lee OO, Chui PY, Wong YH, Pawlik JR, Qian PY (2009) Evidence for vertical transmission of bacterial symbionts from adult to embryo in the Caribbean sponge Svenzea zeai . Appl Environ Microb 75: 6147–6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schmitt S, Angermeier H, Schiller R, Lindquist N, Hentschel U (2008) Molecular microbial diversity survey of sponge reproductive stages and mechanistic insights into vertical transmission of microbial symbionts. Appl Environ Microb 74: 7694–7708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Clark DA, Norris PR (1996) Acidimicrobium ferrooxidans gen nov, sp nov: Mixed-culture ferrous iron oxidation with Sulfobacillus species. Microbiol UK 142: 785–790. [DOI] [PubMed] [Google Scholar]
- 56. Clum A, Nolan M, Lang E, Del Rio TG, Tice H, et al. (2009) Complete genome sequence of Acidimicrobium ferrooxidans type strain (ICP(T)). Stand Genomic Sci 1: 38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Davis-Belmar CS, Norris PR (2009) Ferrous iron and pyrite oxidation by "Acidithiomicrobium" species. Adv Mater Res 71–73: 271–274. [Google Scholar]
- 58. Johnson DB, Bacelar-Nicolau P, Okibe N, Thomas A, Hallberg KB (2009) Ferrimicrobium acidiphilum gen. nov., sp. nov. and Ferrithrix thermotolerans gen. nov., sp. nov.: heterotrophic, iron-oxidizing, extremely acidophilic Actinobacteria . Int J Syst Evol Micr 59: 1082–1089. [DOI] [PubMed] [Google Scholar]
- 59.Imhoff JF (2005) Order I. Chromatiales ord. nov. In: Brenner DJ, Krieg NR, Staley NR, Garrity GM, editors. Bergeýs Manual of Systematic Bacteriology, 2nd edn, vol. 2 (The Proteobacteria), part B (The Gammaproteobacteria). New York: Springer. 1–3.
- 60. Proctor LM (1997) Nitrogen-fixing, photosynthetic, anaerobic bacteria associated with pelagic copepods. Aquat Microb Ecol 12: 105–113. [Google Scholar]
- 61. Schmitt S, Deines P, Behnam F, Wagner M, Taylor MW (2011) Chloroflexi bacteria are more diverse, abundant, and similar in high than in low microbial abundance sponges. FEMS Microbiol Ecol 78: 497–510. [DOI] [PubMed] [Google Scholar]
- 62. Bruck WM, Bruck TB, Self WT, Reed JK, Nitecki SS, et al. (2010) Comparison of the anaerobic microbiota of deep-water Geodia spp. and sandy sediments in the Straits of Florida. ISME J 4: 686–699. [DOI] [PubMed] [Google Scholar]
- 63. Siegl A, Hentschel U (2010) PKS and NRPS gene clusters from microbial symbiont cells of marine sponges by whole genome amplification. Environ Microbiol Rep 2: 507–513. [DOI] [PubMed] [Google Scholar]
- 64.Garrity (2006) Order III. Rhodobacterales ord. nov. In: Brenner DJ, Krieg NR, Staley NR, Garrity GM, editors. Bergeýs Manual of systematic Bacteriology, 2nd edn, vol. 2 (The Proteobacteria), part C (The Alphaproteobacteria, Betaproteobacteria, Deltaproteobacteria and Epsilonproteobacteria). New York: Springer. 161–167.
- 65. Hiraishi A, Ueda Y (1995) Isolation and characterization of Rhodovulum strictum sp. nov. and some other purple nonsulfur bacteria from colored blooms in tidal and seawater pools. Int J Syst Bacteriol 45: 319–326. [DOI] [PubMed] [Google Scholar]
- 66. Srinivas T, Kumar PA, Sasikala C, Ramana CV (2007) Rhodovulum imhoffii sp. nov. Int J Syst Evol Micr 57: 228–232. [DOI] [PubMed] [Google Scholar]
- 67.Imhoff JF (2005) Genus XIV. Rhodovulum Hiraishi and Ueda 1994. In: Brenner DJ, Krieg NR, Staley NR, Garrity GM, editors. Bergeýs Manual of systematic Bacteriology, 2nd edn, vol. 2 (The Proteobacteria), part C (The Alphaproteobacteria, Betaproteobacteria, Deltaproteobacteria and Epsilonproteobacteria),. New York: Springer. 205–209.
- 68.Brusca RC, Brusca GJ (2002) Phylum Porifera: the sponges,. In Sunauer AD, editor. Invertebrates. Sinauer Associates, Inc.,Cambridge, MA. 179–208.
- 69. Hentschel U, Hopke J, Horn M, Friedrich AB, Wagner M, et al. (2002) Molecular evidence for a uniform microbial community in sponges from different oceans. Appl Environ Microb 68: 4431–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. McLachlan J (1964) Some considerations of growth of marine algae in artificial media. Can J Microbiol 10: 769–772. [DOI] [PubMed] [Google Scholar]
- 71. Meyer CP, Geller JB, Paulay G (2005) Fine scale endemism on coral reefs: Archipelagic differentiation in turbinid gastropods. Evolution 59: 113–125. [PubMed] [Google Scholar]
- 72. Folmer O, Black M, Hoeh W, Lutz R Vrijenhoek. R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotech 3: 294–299. [PubMed] [Google Scholar]
- 73. Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Garson MJ, Flowers AE, Webb RI, Charan RD, McCaffrey EJ (1998) A sponge/dinoflagellate association in the haplosclerid sponge Haliclona sp.: cellular origin of cytotoxic alkaloids by Percoll density gradient fractionation. Cell Tissue Res 293: 365–373. [DOI] [PubMed] [Google Scholar]
- 75. Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173: 697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Heuer H, Krsek M, Baker P, Smalla K, Wellington EMH (1997) Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl Environ Microb 63: 3233–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Costa R, Götz M, Mrotzek N, Lottmann J, Berg G, et al. (2006) Effects of site and plant species on rhizosphere community structure as revealed by molecular analysis of microbial guilds. FEMS Microbiol Ecol 56: 236–249. [DOI] [PubMed] [Google Scholar]
- 78. Gomes NCM, Heuer H, Schonfeld J, Costa R, Mendonça -Hagler L, et al. (2001) Bacterial diversity of the rhizosphere of maize (Zea mays) grown in tropical soil studied by temperature gradient gel electrophoresis. Plant Soil 232: 167–180. [Google Scholar]
- 79.Rademaker J, de Bruijn FJ (2004) Computer-assisted analysis of molecular fingerprint profiles and database construction, In: GA Kowalchuk, FJ de Bruijn, IM Head, AD Akkermans, JD van Elsas editors. Molecular Microbial Ecology Manual. Kluwer Academic Publishers, Dordrecht, The Netherlands. 1397–1446.
- 80. Costa R, Salles JF, Berg G, Smalla K (2006) Cultivation-independent analysis of Pseudomonas species in soil and in the rhizosphere of field-grown Verticillium dahliae host plants. Environ Microbiol 8: 2136–2149. [DOI] [PubMed] [Google Scholar]
- 81. Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig WG, et al. (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188–7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ludwig W, Strunk O, Westram R, Richter L, Meier H, et al. (2004) ARB: a software environment for sequence data. Nucleic Acids Res 32: 1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hardoim CCP, Cox CJ, Peixoto RS, Rosado AS, Costa R, et al. (2012) Diversity of the candidate phylum Poribacteria in the marine sponge Aplysina fulva. Braz J Microbiol. In press. [DOI] [PMC free article] [PubMed]
- 85. Keane TM, Creevey CJ, Pentony MM, Naughton TJ, McInerney JO (2006) Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol 6: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Rodriguez F, Oliver JL, Marin A, Medina JR (1990) The general stochastic-model of nucleotide substitution. J Theor Biol 142: 485–501. [DOI] [PubMed] [Google Scholar]
- 87. Huelsenbeck JP, Ronquist F (2001) MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- 88. Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 89. Stamatakis A (2006) RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sponge species. In situ pictures of S. spinosulus (A) and I. variabilis (B).
(TIF)
Cluster analysis. Cluster analysis of PCR-DGGE fingerprints obtained for Bacteria (A), Actinobacteria (B) and Alphaproteobacteria (C). S. spinosulus: Alg10/08, Alg10/09, Alg10/10 and Alg10/11; I. variabilis: Alg10/12, Alg10/13, Alg10/14 and Alg10/15 and Seawater: SW07, SW22, SW23 and SW24.
(TIF)
PCR-DGGE band richness, diversity and evenness.
(DOCX)