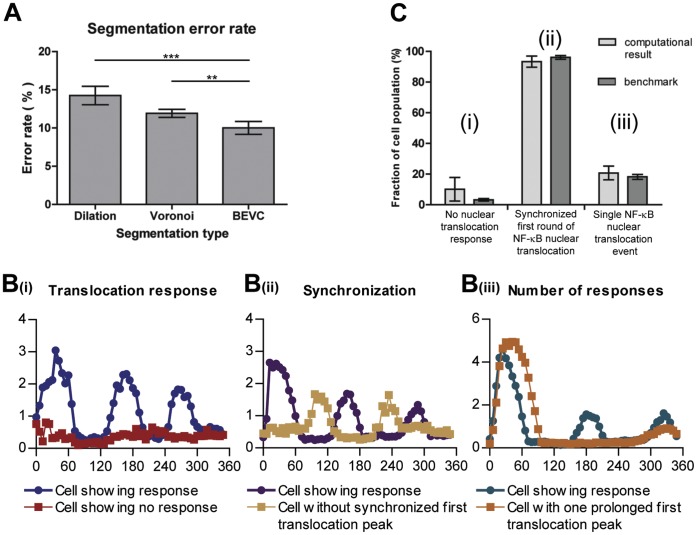
Figure 4. Statistical validation of the automated image segmentation and NF-κB translocation quantification.
(A) Comparison of 3 cytoplasmic segmentation methods based on the criterion of error rate. The error rate of the Dilation method is 14.5%±3.2; of Voronoi it is 11.8%±1.4; and of BEVC it is 10.3%±2.2.* p-value<0.05; ** p-value <0.005; Paired t test (B) Example translocation profiles of (i) cells without translocation and cells with translocation, (ii) cells with and without a synchronized first round of NF-κB translocation, (iii) cells with NF-κB translocation occurring only once and cells with more than one NF-κB translocation event. (C) Bias assessment of our quantification method by comparison of the computational results with the benchmark for different subpopulation. No significant differences (P-value >0.1) were found between the computational results and the benchmark for the different cell subpopulations within a 6 hour imaging timeframe.