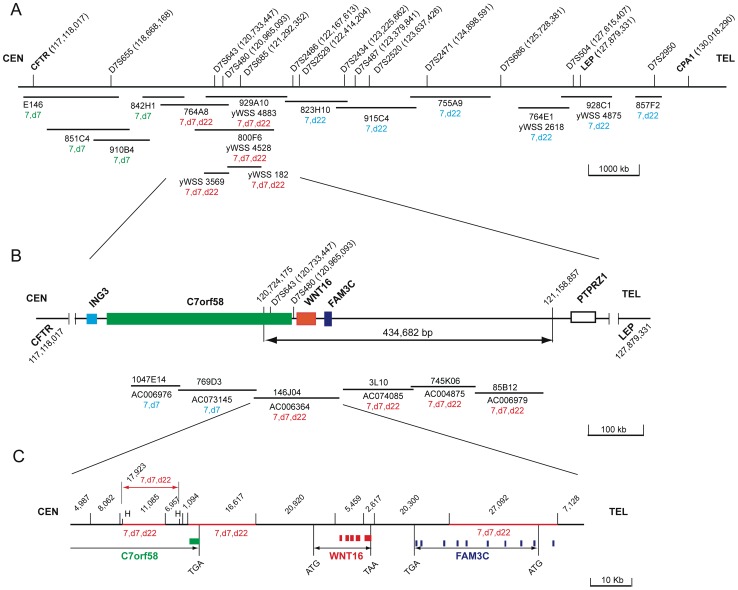
Figure 4. FISH maps of YAC and BAC DNA clones from chromosome 7q31.1 in and around the duplicated region.
The hg19 genomic coordinates of each marker and gene are indicated next to its designated name. The centromeric (CEN) and telomeric (TEL) directions are specified for orientation on the chromosome. Clones are denoted by their clone names, plate coordinates or/and Genbank designations. Each clone or restriction fragment is also denoted by its hybridization onto the patient’s chromosomes 7, der(7) and/or der(22). (A) FISH map of a 12.9 Mb region of chromosome 7 extending from the CFTR to the CPA1 genes. The denoted YAC probes represent three distinct areas of hybridization relative to the chromosome 7 translocation breakpoint, namely distal (7, der22), at or around the breakpoint (7, der7, der22) and proximal to the breakpoint (7, der7). (B) FISH map of BAC clones at and proximal to the 434,682 bp duplicated region, spanning the tail end of C7orf58 and the entire WNT16 and FAM3C genes. Note that all BAC clones within the duplicated region map to 7,d7,d22. (C) Bam HI restriction map, except for two designated Hind III (H) sites, of BAC 146J04 showing the FISH mapping of Bam HI and Hind III restriction fragments, indicating hybridization to 7,der(7) and der(22). The sizes in bp of each restriction fragment are shown and the exons in each gene displayed with solid boxes. The initiation (ATG) and termination (TGA, TAA) codons denote the gene boundaries except for C7orf58, which is only spanned by its last exon and part of the adjacent intron.