Abstract
Background
With development from immature fetus, to near-term fetus, to newborn, to adult, the cerebral vasculature undergoes a number of fundamental changes. Although, the near-term fetus is prepared for a transition from an intra- to extra- uterine existence, this is not necessarily the case with the premature fetus, which is more susceptible to cerebrovascular dysregulation. In the present study, we tested the hypothesis that the profound developmental and age-related differences in cerebral blood flow are associated with significant underlying changes in gene expression.
Methods
With the use of oligonucleotide microarray and pathway analysis, we elucidated significant changes in the transcriptome with development in sheep carotid arteries (CA).
Results
We demonstrate a U-shaped relationship of gene expression during early life, compared to adult for major cerebrovascular network/pathways, e.g. gene expression in the premature fetus and newborn is considerably greater than that of the near-term fetus. Specifically, cell proliferation, growth, and assembly pathway genes were up-regulated during early life. In turn, compared to adult, mitogen activated protein kinase-extracellular regulated kinase, actin cytoskeleton, integrin signaling pathways were down-regulated during early life.
Conclusion
In cerebral vascular smooth muscle, the present studies demonstrated significant changes in important cellular and signaling pathways with maturational development.
Introduction
Neurological impairments such as cerebrovascular accidents and transient ischemic attacks are far too common problems, which increase in prevalence with aging (1). Moreover, in newly born infants hemorrhage into the germinal matrix and periventricular region, occurs in 2 to 5 per 1000 live births and is associated with the development of severe neurological sequelae such as cerebral palsy, convulsive disorders, and other diseases (2). Among very preterm low birth weight (<32 weeks gestation; ≦1500 g) and particularly extremely low birthweight (< 28 weeks gestation; < 1000 g) infants the prevalence of brain damage is particularly high (3). These conditions underscore the importance of well regulated cerebral blood flow (CBF) during perinatal development. Moreover, the transitions which occur at the time of birth, constitute the single most dramatic physiologic event in the life of an individual. In these few moments of parturition, the central circulatory pattern must change from one based on placental transfer of respiratory gases to one of pulmonary ventilation. Systemic vascular resistance increases dramatically, as does arterial blood pressure, while pulmonary vascular resistance and pressure fall. Cardiac (i.e., left ventricular) output initially increases, and then slowly decreases over succeeding days. Despite these dramatic changes in cardiac function and vascular resistance, blood flow to the brain increases only slightly to maintain optimal cerebral oxygenation and metabolism (4). In addition to the cerebral vasculature, per se, carotid arteries (CA) play a crucial role in maintaining optimal CBF (5). Studies have demonstrated a significant pressure gradient from CA to cerebral arteries (6), probably to minimize the exposure of high pressure to delicate cerebral arteries. and underscore the importance of CA in the regulation of CBF. Importantly, studies suggest that much of the change in systemic pressure results in dilation/contraction of the large arteries that supply the brain (7). Therefore, failure of CA to effectively regulate the pressure of the blood reaching delicate cerebral arteries may result in their hemorrhagic rupture. Yet other evidence in premature infants suggests that larger arteries are not able to regulate CBF effectively as in the near-term babies (5).
In general, an infant born at 37 weeks gestation or thereafter, the cerebrovascular physiologic transitions usually occur in a well orchestrated fashion. At younger ages (<37 weeks, preterm; <28 weeks, extremely preterm), however, they may not occur properly with resultant CBF dysregulation (8). In addition to the changes in vascular dynamics, birth also is associated with a large number of major changes in circulating concentrations of various vasoactive hormones and metabolites (9). These include marked increases in norepinephrine and epinephrine, cortisol, the prostaglandins (PGF2α, PGI2, PGD2), angiotensin II, thyroid stimulating hormone, and triiodothyronine, as well as increases in bradykinin, free fatty acids, and glycerol. In contrast, the concentrations of circulating adenosine, growth hormone, and PGE2 decrease dramatically (9). Each of these compounds plays an important role in the regulation of vascular reactivity, as well as circulation to the brain and other organs.
Of importance, the cerebral vasculature also undergoes a number of changes with maturational development. During the past several decades, others and our studies have revealed important aspects, in the fundamental signaling mechanisms that regulate cerebrovascular contractility with maturational development in the fetus and newborn, compared to adult (10, 11). Nonetheless,, the fundamental biochemical and molecular mechanisms responsible for these developmental changes are poorly understood. Some of these important mechanistic differences include: unique features of calcium (Ca2+) -dependent receptor-second messenger coupling with plasma membrane potassium (K+)- and Ca2+-channels, the virtual dependence of the immature organism on extracellular Ca2+ (as opposed to intracellular Ca2+ stores in adult) for Ca2+-dependent thick (myosin) filament regulation (12). Additionally, many elements of the non-Ca2+-dependent pathway of protein kinase C (PKC) to specific enzymes such as extra-cellular regulated kinases (ERK1/2) and their downstream effectors differ in the fetus, compared to adult (13). Taken together, these and other differences account for the significantly greater Ca2+ sensitivity of the cerebrovascular contractile mechanisms of the fetus and newborn, as compared to adult. These studies also emphasize the need to understand the molecular basis of these changes. Unfortunately, and of critical importance, our current understanding of the role of gene expression that underlies cerebrovascular homeostatic mechanisms, during maturational development is extremely limited.
To address this vital issue, by use of oligonucleotide microarrays and signal pathway analysis, we tested the hypothesis that the profound age-related differences in the cerebral artery reactivity are associated with significant underlying changes in the gene expression. We examined changes in gene expression in the carotid arteries from four age groups of sheep: premature fetus, near-term fetus, newborn, and adult.
Methods
Experimental animals and tissues
All experimental procedures were performed within the regulations of the Animal Welfare Act, the National Institutes of Health Guide for the Care and Use of Laboratory Animals, “The Guiding Principles in the Care and Use of Animals” approved by the Council of the American Physiological Society, and the Animal Care and Use Committee of Loma Linda University. For these studies, we used carotid arteries from premature (95 gestational day; 95 GD) fetus, near-term fetus (~140 GD fetus), newborn lamb (1–5 day old) and non-pregnant adult sheep (18–24 months) obtained from Nebeker Ranch (Lancaster, CA). For each experiment 4 animals were used, in case of fetal twins only one of the twins was included in the study.
Pregnant and non-pregnant ewes were anesthetized with thiopental sodium (10 mg/kg, i.v.), and anesthesia was maintained with inhalation of 1% isoflurane in oxygen throughout surgery. Following delivery of the fetus by hysterotomy, the fetuses and ewes were euthanized with an overdose of the proprietary euthanasia solution, Euthasol (pentobarbital sodium 100 mg/Kg and phenytoin sodium 10 mg/Kg; Virbac, Ft. Worth, TX). Studies were performed in isolated carotid arteries cleaned of adipose and connective tissue. To avoid the complications of endothelial-mediated effects, we removed the endothelium by carefully inserting a small wire three times, as previously described (11).
Tissue collection and microarray processing
In previous studies, we have described this technique in detail (14). Microarray analysis was conducted by utilizing commercial services of GenUs Biosystems, Northbrook, Illinois. Briefly, tissue samples were lysed in Tri-reagent (Ambion, Austin, TX) and total RNA was isolated using phenol/chloroform extraction followed by purification over spin columns (Ambion). The concentration and purity of total RNA was measured by spectrophotometry at OD260/280 and the quality of the total RNA sample was assessed using an Agilent Bioanalyzer with the RNA6000 Nano Lab Chip (Agilent Technologies, Santa Clara, CA).
Labeled cRNA was prepared by linear amplification of the Poly(A)+ RNA population within the total RNA sample. Briefly, <1 μg of total RNA was reverse transcribed after priming with a DNA oligonucleotide containing the T7 RNA polymerase promoter 5′ to a d(T)24 sequence. After second-strand cDNA synthesis and purification of double-stranded cDNA, in vitro transcription was performed using T7 RNA polymerase. The quantity and quality of the labeled cRNA was assayed by spectrophotometry and the Agilent Bioanalyzer.
One μg of purified cRNA was fragmented to uniform size and applied to Agilent Sheep Gene Expression Microarray, 8 × 15K (Design ID 019921, Agilent Technologies) in hybridization buffer. Arrays were hybridized at 65° C for 17 hrs. in a shaking incubator and washed at 37° C for 1 min. Rinsed and dried arrays were scanned with an Agilent G2565 Microarray Scanner (Agilent Technologies) at 5 μm resolution. Agilent Feature Extraction software was used to process the scanned images from arrays (gridding and feature intensity extraction) and the data generated for each probe on the array was analyzed with GeneSpring GX v7.3.1 software (Agilent Technologies). Annotations are based on the Agilent eArray annotation file dated January, 2010.
Pathway/Network Analysis
Each gene was annotated manually using NCBI Blast Search, Unigene, Entrez or other databases. We then analyzed the annotated genes using Ingenuity Pathway Analysis Program (Ingenuity Systems, Redwood City, CA).
Real time PCR validation
To validate the results of the microarray analysis, we chose stathmin (STMN1), filamin A alpha (FLNA), and myosin light chain kinase (MYLK) genes that were highly regulated by development during early life, compared to adult, for analysis using real time PCR. Using the same probe sequences as those on the microarray chip, we designed primers with the use of Primer 3 web-based software (http://frodo.wi.mit.edu/primer3/). The primers were synthesized by Integrated DNA technologies (Coralville, CA). Total RNA (1 ug per reaction) was reverse transcribed using Quantitect reverse transcriptase kit (Qiagen, Valencia, CA). Relative expression was normalized to 18S RNA and fold changes were calculated using the ΔΔ cycle threshold (CT) method (15). Samples were analyzed on the Roche LightCycler 1.5 (Roche, Indianapolis, IN).
Statistics
To compare individual expression values across arrays, raw intensity data from each gene was normalized to the 75th percentile intensity of each array. Only genes with values greater than background intensity for all samples within each group were used for further analysis. Differentially expressed genes were identified by 2-fold change and Welch T-test p-values <0.05 between each treatment group and its age-specific normoxic control. Statistical significance in the real-time PCR data was determined by one-way analysis of variance (ANOVA) and post-hoc Newmans-Keul test.
Results
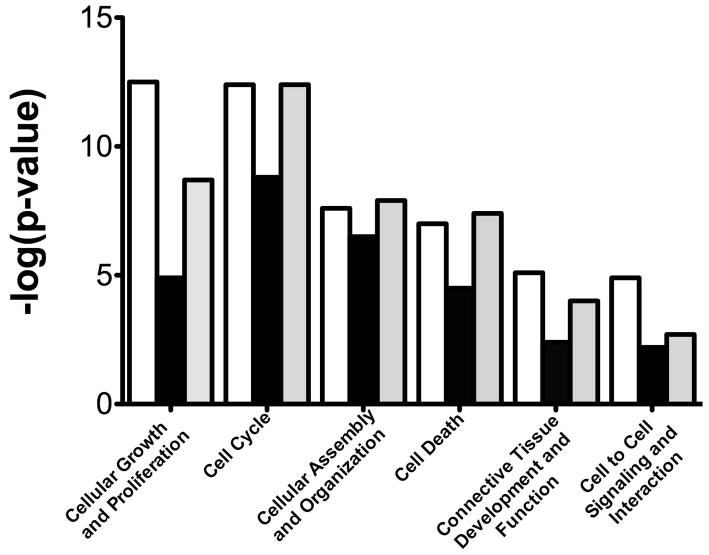
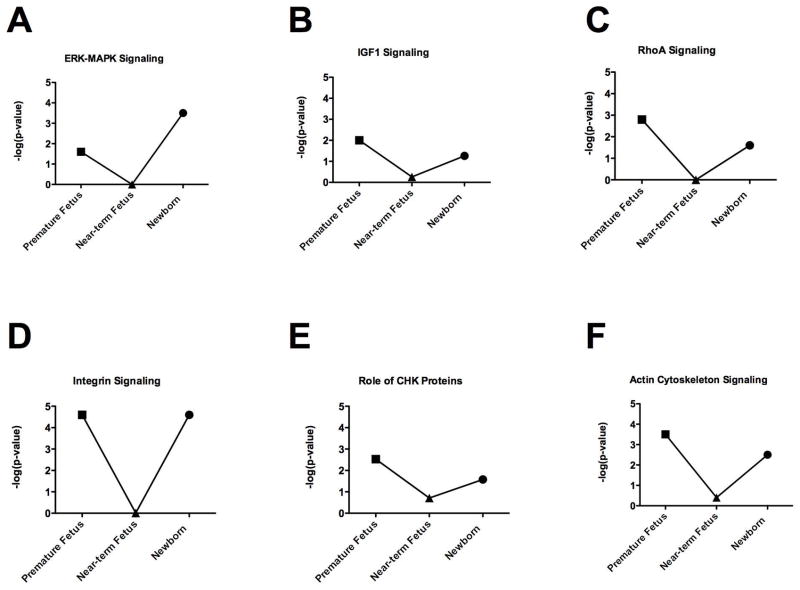
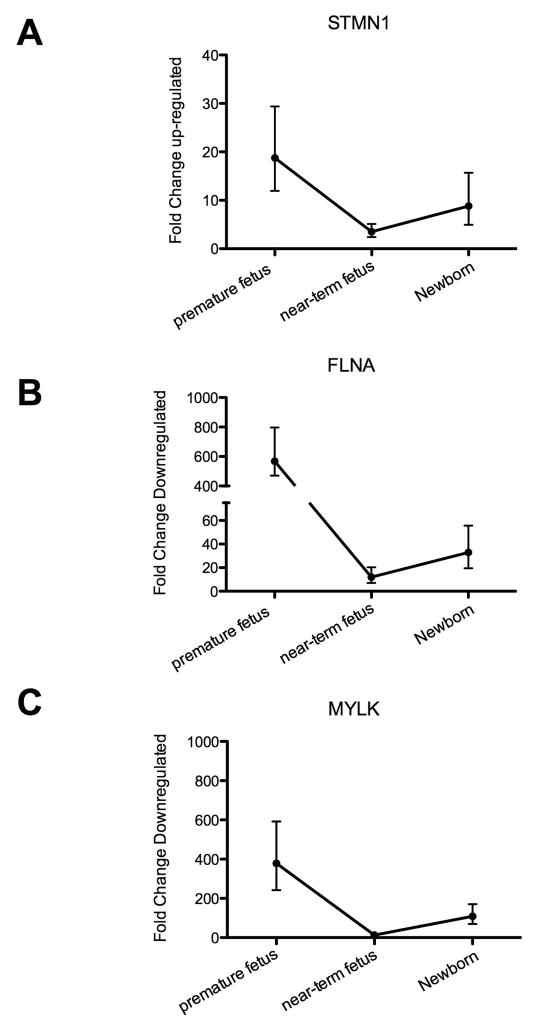
Our results demonstrate profound changes in ovine carotid artery gene expression profiles with developmental maturation from premature fetus to mature fetus, to newborn, to adult. Table 1 enumerates the number of genes with up- and down-regulated expression (both > 2 and >4 fold-change and p-value) in premature fetus, near-term fetus, and newborn lamb compared to adult sheep. In a striking manner, the changes in CA gene expression profiles from premature fetus and newborn lamb, compared to adult, differed to a similar extent. In contrast and compared to adult, in the near-term fetal CA fewer genes showed differential regulation. A similar pattern of increased changes in CA gene expression in premature fetus and newborn lamb with far fewer changes in near-term fetus also was observed in several functional (Figure 1) as well as canonical (Figure 2) gene pathways/networks. Striking is the “U” shaped pattern of these gene expression responses. The main canonical pathways altered in early life (premature fetus, near-term fetus, and newborn) compared to adult were G2/M DNA damage checkpoint regulation, mitotic roles of polo-like kinases, and cyclins and cell cycle regulation pathways. Of relevance, these pathways regulate a number aspects of cellular growth, proliferation, assembly, DNA replication, cell development, maintenance and so forth. To validate the relative protein expression as development proceeds, Figure 3 demonstrates that the CA expression of STMN1, FLNA, and MYLK in from premature fetus, near-term fetus, and newborn follows the trend of the microarray analysis.
Table 1.
Number of Genes Altered with Development
| Premature Fetus | Near-term Fetus | Newborn | |
|---|---|---|---|
| Genes Altered (>2 Fold; P < 0.05) | |||
| Up-regulated | 2570 | 1212 | 2371 |
| Down-regulated | 1907 | 658 | 1512 |
| Genes Altered (> 4 Fold: P < 0.001) | |||
| Up-regulated | 373 | 110 | 304 |
| Down-regulated | 255 | 11 | 122 |
Figure 1.
Functional pathways altered with development. Bar graph demonstrates functional pathways altered with development. N was 4 in each experimental group, and all groups were significantly different compared to adult (P < 0.05). White, black, and grey bars shows comparison of adult with premature fetus, near-term fetus, and newborn, respectively.
Figure 2.
Chief canonical pathways altered with development. Significant differences [−log(p-value)] in the (A) ERK-MAPK, (B) IGF1, (C) Ras Homolog Gene Family Member A (RhoA), (D) Integrin, (E) Role of checkpoint (CHK) proteins, and (F) Actin cytoskeleton signaling pathways in the carotid arteries from premature fetus, near-term fetus, and newborn lamb, compared to adult are shown in a line-graph format. N was 4 in each experimental group, and all groups were significantly different compared to adult (P < 0.05).
Figure 3.
Real-time PCR validation of microarray analysis. Demonstrates changes in the expression of (A) STMN1, (B) FLNA, and (C) MYLK mRNA levels in the carotid arteries from premature fetus, near-term fetus, and newborn lamb, compared to adult as determined by quantitative real-time PCR analysis. N was 4 in each experimental group, and all groups were significantly different compared to adult (P < 0.05).
Tables 2 to 4 enumerate the top 20 genes with up-regulated expression in carotid arteries from premature fetus (Table 2), near-term fetus (Table 3), and newborn lamb (Table 4). Tables 5 to 7 list the top 10 to 20 genes with down-regulated expression in CA from premature fetus (Table 5), near-term fetus (Table 6), and newborn lamb (Table 7), compared to adult sheep. Table 8 lists the major genes involved in cellular growth, proliferation, and assembly pathways that have significantly up-regulated expression during early life. Similarly, Table 9 lists those genes with down-regulated expression in early life, compared to adult. The genes with down-regulated expression belonged to the integrin, actin cytoskeleton, and PKC-Rho Kinase-mitogen activated protein kinase (MAPK) pathways.
Table 2.
Top 20 Up-regulated Genes in Premature Fetus as compared to Adult
| Symbol | Name | Fold Change | P-Value |
|---|---|---|---|
| CTHRC1 | collagen triple helix repeat containing 1 | 454.000 | 6.38E–05 |
| HBB | hemoglobin, beta | 390.200 | 1.03E–05 |
| UBE2C | ubiquitin-conjugating enzyme E2C | 259.300 | 1.89E–07 |
| CRABP1 | cellular retinoic acid binding protein 1 | 245.300 | 2.23E–04 |
| KIAA0101 | KIAA0101 | 211.400 | 3.12E–08 |
| COL21A1 | collagen, type XXI, alpha 1 | 208.100 | 9.59E–06 |
| BIRC5 | baculoviral IAP repeat containing 5 | 177.400 | 6.28E–06 |
| MEST | mesoderm specific transcript homolog | 173.800 | 7.54E–05 |
| ACSM1 | acyl-CoA synthetase medium-chain family member 1 | 114.900 | 8.84E–05 |
| NCAPG | non-SMC condensin I complex, subunit G | 100.500 | 8.00E–07 |
| RBP1 | retinol binding protein 1, cellular | 81.650 | 5.86E–04 |
| CPZ | carboxypeptidase Z | 75.910 | 1.29E–07 |
| CKS2 | CDC28 protein kinase regulatory subunit 2 | 73.800 | 4.67E–08 |
| STMN1 | stathmin 1 | 73.070 | 5.69E–05 |
| TF | transferrin | 65.610 | 3.12E–05 |
| CDCA3 | cell division cycle associated 3 | 64.230 | 2.42E–07 |
| APOA1 | apolipoprotein A-I | 63.910 | 9.65E–04 |
| CENPE | centromere protein E, 312kDa | 61.960 | 1.10E–05 |
| CDCA7 | cell division cycle associated 7 | 61.590 | 4.44E–06 |
| AURKB | aurora kinase B | 58.150 | 4.34E–07 |
Table 4.
Top 20 Up-regulated Genes in newborn as compared to Adult
| Symbol | Name | Fold Change | P-Value |
|---|---|---|---|
| UBE2C | ubiquitin-conjugating enzyme E2C | 146.500 | 3.72E–07 |
| ACSM1 | acyl-CoA synthetase medium-chain family member 1 | 130.000 | 7.60E–04 |
| IAA0101 | KIAA0101 | 106.400 | 3.88E–05 |
| COL21A1 | collagen, type XXI, alpha 1 | 95.850 | 1.00E–03 |
| BIRC5 | baculoviral IAP repeat containing 5 | 82.050 | 1.81E–05 |
| NCAPG | non-SMC condensin I complex, subunit G | 74.720 | 1.24E–06 |
| OGN | osteoglycin | 73.130 | 9.99E–08 |
| HAPLN1 | hyaluronan and proteoglycan link protein 1 | 72.410 | 8.26E–06 |
| CKS2 | CDC28 protein kinase regulatory subunit 2 | 57.910 | 3.18E–06 |
| DIAPH3 | diaphanous homolog 3 (Drosophila) | 46.560 | 9.83E–07 |
| CENPE | centromere protein E, 312kDa | 42.730 | 3.05E–06 |
| CDK1 | cyclin-dependent kinase 1 | 41.470 | 1.14E–05 |
| GPX8 | glutathione peroxidase 8 (putative) | 39.530 | 3.04E–05 |
| CCNA2 | cyclin A2 | 39.310 | 7.80E–06 |
| MEM45A | transmembrane protein 45A | 35.950 | 5.09E–05 |
| APOC2 | apolipoprotein C-II | 35.900 | 5.67E–04 |
| LOX | lysyl oxidase | 35.710 | 4.65E–05 |
| PRDX4 | peroxiredoxin 4 | 34.720 | 2.39E–05 |
| ORF4L1 | mortality factor 4 like 1 | 32.890 | 8.05E–04 |
| MEST | mesoderm specific transcript homolog (mouse) | 28.330 | 1.04E–04 |
Table 3.
Top 20 Up-regulated Genes in near-term Fetus as compared to Adult
| Symbol | Name | Fold Change | P-Value |
|---|---|---|---|
| ALDOA | aldolase A, fructose-bisphosphate | 67.02 | 2.62E–04 |
| UBE2C | ubiquitin-conjugating enzyme E2C | 66.4 | 7.52E–05 |
| MEG3 | maternally expressed 3 (non-protein coding) | 63.3 | 3.19E–06 |
| ELN | elastin | 55.59 | 3.90E–04 |
| CRABP1 | cellular retinoic acid binding protein 1 | 51.01 | 6.76E–04 |
| MEST | mesoderm specific transcript homolog (mouse) | 50.56 | 5.57E–05 |
| APOC2 | apolipoprotein C-II | 50.26 | 7.34E–05 |
| NCAPG | non-SMC condensin I complex, subunit G | 46.93 | 6.68E–06 |
| HBB | hemoglobin, beta | 43.91 | 9.42E–06 |
| CENPE | centromere protein E, 312kDa | 42.58 | 9.18E–06 |
| MFAP2 | microfibrillar-associated protein 2 | 38.33 | 1.83E–04 |
| CDCA3 | cell division cycle associated 3 | 37.42 | 1.32E–05 |
| BIRC5 | baculoviral IAP repeat containing 5 | 33.53 | 2.65E–04 |
| STAB1 | stabilin 1 | 33.1 | 6.00E–04 |
| FST | follistatin | 32.86 | 2.39E–04 |
| FBLN7 | fibulin 7 | 32.35 | 1.31E–04 |
| RNASE1 | ribonuclease, RNase A family, 1 (pancreatic) | 32.34 | 6.37E–04 |
| CKS2 | CDC28 protein kinase regulatory subunit 2 | 30.33 | 1.50E–06 |
| AURKB | aurora kinase B | 30.03 | 3.66E–06 |
| HAPLN1 | hyaluronan and proteoglycan link protein 1 | 29 | 5.78E–05 |
Table 5.
Top down-regulated Genes in Premature Fetus as compared to Adult
| Symbol | Name | Fold Change | P-Value |
|---|---|---|---|
| DES | desmin | −132.979 | 1.56E–04 |
| MUSTN1 | musculoskeletal, embryonic nuclear protein 1 | −102.459 | 9.41E–04 |
| ITGA5 | integrin, alpha 5 (fibronectin receptor, alpha polypeptide) | −39.062 | 7.94E–07 |
| SOD3 | superoxide dismutase 3, extracellular | −33.003 | 1.05E–04 |
| PPP1R15A | protein phosphatase 1, regulatory (inhibitor) subunit 15A | −29.326 | 5.16E–05 |
| CSRP1 | cysteine and glycine-rich protein 1 | −28.249 | 2.72E–08 |
| PACS1 | phosphofurin acidic cluster sorting protein 1 | −26.738 | 2.45E–07 |
| FBXO32 | F-box protein 32 | −24.450 | 1.75E–04 |
| ZYX | zyxin | −22.936 | 3.75E–04 |
| MID1IP1 | MID1 interacting protein 1 (gastrulation specific G12 homolog | −21.598 | 1.53E–07 |
| ZFP36 | zinc finger protein 36, C3H type, homolog | −18.904 | 7.75E–04 |
| MYL9 | myosin, light chain 9, regulatory | −18.832 | 1.80E–06 |
| UNC45A | unc-45 homolog A (C. elegans) | −17.953 | 6.82E–05 |
| SLC2A4 | solute carrier family 2 (facilitated glucose transporter), member 4 | −17.422 | 3.47E–05 |
| FLNA | filamin A, alpha | −16.807 | 4.21E–06 |
| CEBPB | CCAAT/enhancer binding protein (C/EBP), beta | −16.529 | 3.33E–05 |
| SPHK1 | sphingosine kinase 1 | −15.974 | 5.99E–04 |
| COPS7A | COP9 constitutive photomorphogenic homolog subunit 7A | −15.674 | 1.34E–06 |
| FBXW5 | F-box and WD repeat domain containing 5 | −15.152 | 5.35E–06 |
| DNAJB5 | DnaJ (Hsp40) homolog, subfamily B, member 5 | −14.006 | 3.06E–07 |
Table 7.
Top down-regulated Genes in Newborn Lamb as compared to Adult
| Symbol | Name | Fold Change | P-Value |
|---|---|---|---|
| PACS1 | phosphofurin acidic cluster sorting protein 1 | −22.727 | 7.20E–07 |
| SOD3 | superoxide dismutase 3, extracellular | −20.877 | 1.10E–04 |
| PCIF1 | PDX1 C-terminal inhibiting factor 1 | −15.625 | 1.67E–06 |
| WBP2 | WW domain binding protein 2 | −15.480 | 1.66E–05 |
| BCL6 | B-cell CLL/lymphoma 6 | −15.038 | 7.42E–04 |
| COPS7A | COP9 constitutive photomorphogenic homolog subunit 7A | −14.837 | 1.29E–06 |
| SCAF1 | SR-related CTD-associated factor 1 | −14.184 | 1.14E–04 |
| SF3A2 | splicing factor 3a, subunit 2, 66kDa | −13.441 | 9.22E–08 |
| ITGA5 | integrin, alpha 5 (fibronectin receptor, alpha polypeptide) | −13.405 | 9.38E–05 |
| ZYX | zyxin | −13.175 | 1.43E–04 |
| EHD2 | EH-domain containing 2 | −12.315 | 1.24E–06 |
| PDAP1 | PDGFA associated protein 1 | −11.792 | 3.62E–06 |
| RHOG | ras homolog gene family, member G (rho G) | −11.507 | 2.06E–04 |
| EFHD2 | EF-hand domain family, member D2 | −10.941 | 5.35E–05 |
| FAM113B | family with sequence similarity 113, member B | −10.471 | 1.58E–04 |
| SORBS3 | sorbin and SH3 domain containing 3 | −10.020 | 1.68E–05 |
| CALCOCO1 | calcium binding and coiled-coil domain 1 | −9.434 | 3.89E–06 |
| FBXW5 | F-box and WD repeat domain containing 5 | −9.434 | 3.22E–06 |
| INO80B | INO80 complex subunit B | −9.434 | 2.72E–05 |
| MID1IP1 | MID1 interacting protein 1 (gastrulation specific G12 homolog) | −9.434 | 2.01E–04 |
Table 6.
Top down-regulated Genes in near-term Fetus as compared to Adult
| Symbol | Name | Fold Change | P-Value |
|---|---|---|---|
| MAPRE2 | microtubule-associated protein, RP/EB family, member 2 | −8.475 | 6.97E–04 |
| HSPA1A/HSPA1B | heat shock 70kDa protein 1A | −6.944 | 7.22E–04 |
| DNAJB5 | DnaJ (Hsp40) homolog, subfamily B, member 5 | −6.711 | 4.54E–04 |
| CAMK2G | calcium/calmodulin-dependent protein kinase II gamma | −5.747 | 4.65E–04 |
| MYOT | myotilin | −5.128 | 4.70E–04 |
| PRKCD | protein kinase C, delta | −4.950 | 8.82E–05 |
| TBC1D1 | TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 | −4.274 | 7.36E–05 |
| CLIP1 | CAP-GLY domain containing linker protein 1 | −4.184 | 3.77E–06 |
| BZW2 | basic leucine zipper and W2 domains 2 | −4.167 | 4.04E–04 |
| SQSTM1 | sequestosome 1 | −4.065 | 8.68E–04 |
Table 8.
Cellular Proliferation, Growth, and Assembly Pathway was up-regulated in early life
| Symbol | Entrez Gene Name | Premature fetus Vs Adult | Near-term fetus Vs Adult | Newborn Vs Adult | |||
|---|---|---|---|---|---|---|---|
| Fold Change | p-value | Fold Change | p-value | Fold Change | p-value | ||
| ASPM | asp (abnormal spindle) homolog, microcephaly associated | 37.38 | 9.41E-06 | 19.21 | 2.04E-05 | 16.29 | 5.45E-05 |
| AURKA | aurora kinase A | 11.78 | 9.23E-05 | 5.694 | 3.68E-04 | 8.833 | 1.46E-04 |
| AURKB | aurora kinase B | 58.15 | 4.34E-07 | 30.03 | 3.66E-06 | 27.24 | 1.35E-04 |
| BIRC5 | baculoviral IAP repeat containing 5 | 177.4 | 6.28E-06 | 33.53 | 2.65E-04 | 82.05 | 1.81E-05 |
| CCNA2 | cyclin A2 | 54.41 | 6.46E-06 | 21.72 | 1.93E-05 | 39.31 | 7.80E-06 |
| CCNB2 | cyclin B2 | 29.65 | 2.08E-07 | 11.42 | 1.67E-04 | 19.98 | 2.42E-05 |
| CDK1 | cyclin-dependent kinase 1 | 51.19 | 1.61E-04 | 15.16 | 1.97E-04 | 41.47 | 1.14E-05 |
| CDK2 | cyclin-dependent kinase 2 | 5.852 | 2.12E–05 | 5.064 | 3.75E–03 | 3.454 | 1.71E–04 |
| CDT1 | chromatin licensing and DNA replication factor 1 | 7.823 | 1.16E–04 | 11.94 | 1.01E–02 | 3.098 | 7.26E–03 |
| CHEK2 | CHK2 checkpoint homolog (S. pombe) | 8.841 | 3.65E–05 | 3.24 | 1.27E–01 | 5.385 | 1.81E–04 |
| CENPE | centromere protein E, 312kDa | 61.96 | 1.10E-05 | 42.58 | 9.18E-06 | 42.73 | 3.05E-06 |
| CENPH | centromere protein H | 12.61 | 3.67E-06 | 5.277 | 1.21E-04 | 7.478 | 9.92E-06 |
| CKAP2 | cytoskeleton associated protein 2 | 30.63 | 1.84E-04 | 19.23 | 1.88E-04 | 19.94 | 8.83E-05 |
| CKS2 | CDC28 protein kinase regulatory subunit 2 | 73.8 | 4.67E-08 | 30.33 | 1.50E-06 | 57.91 | 3.18E-06 |
| DIAPH3 | diaphanous homolog 3 (Drosophila) | 35.37 | 4.90E-06 | 27.37 | 1.28E-06 | 46.56 | 9.83E-07 |
| ESPL1 | extra spindle pole bodies homolog 1 (S. cerevisiae) | 28.7 | 1.49E-04 | 12.95 | 7.42E-05 | 15.62 | 2.57E-05 |
| GATA6 | GATA binding protein 6 | 33.72 | 8.69E-05 | 8.271 | 2.27E-04 | 16.16 | 1.27E-05 |
| KNTC1 | kinetochore associated 1 | 25.58 | 2.16E-04 | 9.572 | 0.00145 | 14.78 | 5.03E-04 |
| MXD3 | MAX dimerization protein 3 | 31.4 | 4.21E-06 | 26.09 | 3.17E-06 | 14.69 | 8.09E-05 |
| NCAPG | non-SMC condensin I complex, subunit G | 100.5 | 8.00E-07 | 46.93 | 6.68E-06 | 74.72 | 1.24E-06 |
| PRC1 | protein regulator of cytokinesis 1 | 23.2 | 5.61E-05 | 21.77 | 5.18E-04 | 17.77 | 5.21E-05 |
| PCNA | proliferating cell nuclear antigen | 10.78 | 1.03E–04 | 3.439 | 4.35E–04 | 9.182 | 1.43E–05 |
| RFC1 | replication factor C (activator 1) 1, 145kDa | 2.064 | 4.06E–03 | 1.066 | 6.71E–01 | 2.092 | 3.06E–03 |
| RFC3 | replication factor C (activator 1) 3, 38kDa | 4.476 | 8.78E–04 | 1.371 | 1.98E–01 | 3.577 | 4.97E–04 |
| RFC4 | replication factor C (activator 1) 4, 37kDa | 2.799 | 1.31E–02 | 1.198 | 7.25E–01 | 1.878 | 6.80E–02 |
| RFC5 | replication factor C (activator 1) 5, 36.5kDa | 2.486 | 2.22E–07 | 1.348 | 1.68E–02 | 2.065 | 1.55E–03 |
| SKA1 | spindle and kinetochore associated complex subunit 1 | 26.35 | 3.44E-06 | 12.14 | 6.36E-06 | 17.75 | 7.18E-05 |
| SOX9 | SRY (sex determining region Y)-box 9 | 8.645 | 2.50E-05 | 9.4 | 0.0017 | 8.419 | 3.40E-04 |
| STMN1 | stathmin 1 | 73.07 | 5.69E-05 | 20.7 | 9.17E-04 | 27.51 | 2.71E-05 |
| TYMS | thymidylate synthetase | 15.49 | 6.62E-06 | 11.37 | 4.44E-05 | 7.212 | 5.09E-05 |
| UBE2C | ubiquitin-conjugating enzyme E2C | 259.3 | 1.89E-07 | 66.4 | 7.52E-05 | 146.5 | 3.72E-07 |
Table 9.
Top three down-regulated Pathways
| Symbol | Entrez Gene Name | Premature fetus Vs Adult | Near-term fetus Vs Adult | Newborn Vs Adult | |||
|---|---|---|---|---|---|---|---|
| Fold Change | p-value | Fold Change | p-value | Fold Change | p-value | ||
| Integrin Signaling Pathway | |||||||
| CAPN1 | calpain 1, (mu/I) large subunit | −4.608 | 1.29E–04 | −1.131 | 5.36E–01 | −5.263 | 8.63E–05 |
| FNBP1 | formin binding protein 1 | −7.143 | 4.46E–05 | −1.543 | 5.59E–02 | −4.000 | 1.57E–04 |
| ITGA5 | integrin, alpha 5 (fibronectin receptor, alpha polypeptide) | −39.062 | 7.94E–07 | −6.579 | 8.73E–03 | −13.405 | 9.38E–05 |
| ITGA8 | integrin, alpha 8 | −12.225 | 1.20E–04 | −2.410 | 1.54E–02 | −2.320 | 3.95E–02 |
| TLN1 | talin 1 | −12.469 | 3.97E–05 | −2.070 | 7.29E–02 | −7.143 | 1.41E–04 |
| VASP | vasodilator-stimulated phosphoprotein | −8.197 | 1.02E–03 | −5.155 | 8.11E–02 | −3.817 | 2.87E–03 |
| ZYX | zyxin | −22.936 | 3.75E–04 | −2.625 | 5.61E–02 | −13.175 | 1.43E–04 |
| Actin Cytoskeleton Pathway | |||||||
| ACTA2 | actin, alpha 2, smooth muscle, aorta | −4.292 | 4.03E–04 | −1.168 | 5.14E–01 | −2.114 | 4.97E–03 |
| ACTC1 | actin, alpha, cardiac muscle 1 | −23.529 | 1.40E–03 | −3.584 | 6.02E–02 | −6.944 | 1.03E–02 |
| ACTG2 | actin, gamma 2, smooth muscle, enteric | −5.291 | 1.37E–03 | −2.347 | 9.19E–03 | −2.387 | 1.24E–02 |
| ACTN1 | actinin, alpha 1 | −11.274 | 1.46E–04 | −3.030 | 1.35E–03 | −7.634 | 3.48E–05 |
| ACTN3 | actinin, alpha 3 | −5.525 | 1.83E–03 | −6.173 | 1.43E–01 | −3.226 | 4.49E–03 |
| ACTR2 | ARP2 actin-related protein 2 homolog (yeast) | −4.255 | 7.40E–05 | −2.809 | 2.69E–02 | −2.188 | 8.49E–04 |
| CFL1 | cofilin 1 (non-muscle) | −6.536 | 5.74E–04 | −1.314 | 5.58E–01 | −6.536 | 1.11E–04 |
| FLNA | filamin A, alpha | −16.807 | 4.21E–06 | −2.577 | 1.28E–02 | −5.587 | 3.82E–05 |
| FNBP1 | formin binding protein 1 | −7.143 | 4.46E–05 | −1.543 | 5.59E–02 | −4.000 | 1.57E–04 |
| GSK3A | glycogen synthase kinase 3 alpha | −8.772 | 6.13E–05 | −1.916 | 1.33E–02 | −6.061 | 1.49E–03 |
| WASF2 | WAS protein family, member 2 | −4.386 | 8.08E–06 | −3.774 | 8.74E–03 | −3.413 | 2.60E–04 |
| TESK1 | testis-specific kinase 1 | −7.463 | 2.31E–03 | −3.922 | 2.25E–01 | −4.926 | 1.12E–02 |
| TGFB1I1 | transforming growth factor beta 1 induced transcript 1 | −9.524 | 2.50E–04 | −2.227 | 9.38E–03 | −6.098 | 6.52E–05 |
| PKC-ERK-ROCK Pathway | |||||||
| PLCB2 | phospholipase C, beta 2 | −5.714 | 4.01E–04 | −1.456 | 3.35E–01 | −3.175 | 7.21E–04 |
| PLCD1 | phospholipase C, delta 1 | −9.259 | 3.64E–03 | −7.463 | 1.73E–02 | −4.367 | 3.41E–05 |
| PIK3CD | phosphoinositide-3-kinase, catalytic, delta polypeptide | −8.264 | 4.17E–06 | −5.319 | 1.01E–01 | −6.452 | 2.43E–04 |
| PRKACA | protein kinase, cAMP-dependent, catalytic, alpha | −7.463 | 2.59E–05 | −1.546 | 1.86E–01 | −6.211 | 6.85E–05 |
| PRKAG2 | protein kinase, AMP-activated, gamma 2 non-catalytic subunit | −4.695 | 6.43E–05 | −3.125 | 9.67E–04 | −2.809 | 8.77E–04 |
| PRKCD | protein kinase C, delta | −8.850 | 4.45E–06 | −4.950 | 8.82E–05 | −3.937 | 4.98E–05 |
| CAMK2G | calcium/calmodulin-dependent protein kinase II gamma | −12.516 | 2.99E–07 | −5.747 | 4.65E–04 | −5.376 | 2.92E–05 |
| ITPR1 | inositol 1,4,5-triphosphate receptor, type 1 | −4.831 | 1.66E–02 | 1.567 | 3.27E–01 | −4.484 | 1.77E–02 |
| PPP1R11 | protein phosphatase 1, regulatory (inhibitor) subunit 11 | −5.155 | 1.82E–04 | −1.299 | 1.09E–01 | −5.747 | 2.55E–04 |
| CAPN1 | calpain 1, (mu/I) large subunit | −4.608 | 1.29E–04 | −1.131 | 5.36E–01 | −5.263 | 8.63E–05 |
| ARHGAP1 | Rho GTPase activating protein 1 | −9.346 | 1.30E–04 | −2.545 | 6.02E–02 | −6.061 | 3.72E–04 |
| RHOB | ras homolog gene family, member B | −5.128 | 6.26E–03 | −1.340 | 4.00E–01 | −4.630 | 6.78E–03 |
| RHOG | ras homolog gene family, member G (rho G) | −5.747 | 9.13E–05 | −1.366 | 2.46E–01 | −11.507 | 2.06E–04 |
| RRAS | related RAS viral (r-ras) oncogene homolog | −13.423 | 9.08E–06 | −2.457 | 1.55E–02 | −5.848 | 1.06E–05 |
| MAP2K2 | mitogen-activated protein kinase kinase 2 | −3.436 | 1.33E–04 | −1.499 | 3.18E–02 | −3.876 | 3.15E–06 |
| MAPK3 | mitogen-activated protein kinase 3 | −3.846 | 4.37E–03 | −2.421 | 3.23E–02 | −5.076 | 5.44E–04 |
| MKNK1 | MAP kinase interacting serine/threonine kinase 1 | −2.183 | 1.55E–04 | −1.715 | 4.39E–02 | −2.331 | 1.09E–04 |
| MKNK2 | MAP kinase interacting serine/threonine kinase 2 | −3.906 | 7.11E–04 | −1.653 | 1.03E–01 | −3.390 | 2.19E–03 |
| MYL9 | myosin, light chain 9, regulatory | −18.832 | 1.80E–06 | −3.003 | 2.30E–02 | −5.181 | 4.87E–05 |
| MYLK | myosin light chain kinase | −11.050 | 2.61E–05 | −3.155 | 9.59E–02 | −3.030 | 1.78E–03 |
Discussion
The present study demonstrates important changes in gene regulation in ovine carotid arteries with developmental maturation from preterm fetus, to term fetus, and to newborn, in comparison to those genes in the adult. The changes in gene expression profiles (both up- and down-regulation) follow a U-shaped pattern from preterm fetus, to near-term fetus, to newborn. The deviations of gene expression (either up- or down-regulated) are much greater in premature fetus and newborn lamb than in the near-term fetus. We are not aware of any such report demonstrating that the changes in gene expression profiles are less in a near-term fetus than those in the premature fetus or newborn. These findings underscore the immense changes in gene expression, which occur during the perinatal period, birth, and the newborn and how these differ in unique and unexpected manner. Of critical importance, several parameters associated with cerebral blood flow such as pH, PCO2, Hemoglobin g%, heart rate, follow a similar U - shaped pattern during preterm, near-term and newborn life (16). Thus, perhaps it should be no surprise that the gene expression follows a similar pattern. Moreever, at present the importance of this finding or its meaning in a deep sense are not clear. Furthermore, similar to the present finding of a U-shaped curve of gene expression during early life, a number of other significant events, such as complete de-methylation and re-methylation of genome, transcriptional silencing of ovum, histones acetylation of spermatic DNA, still need explanation and the present findings add to this list.
The present study demonstrates that collagen triple helix repeat containing protein 1 (Cthrc1) was up-regulated ~ 450-fold in premature fetus (Table 3); however, its expression decreased 47-fold in near-term fetus and 35-fold in newborn cerebral arteries. Cthrc1 is a gene product with novel biochemical activities, and its ability to reduce collagen deposition by inhibition of Smad2/3 activation plays a major role in vascular development, repair, and fibrosis (17). Of importance, Cthrc1 also increases cellular migration and reduces collagen deposition (18). Recently, we reported that from an ultrastructural standpoint cerebral arteries of the premature fetus are significantly more fragile than those of the near-term fetus (19). Cthrc1 up-regulation may be responsible for reduced collagen contents and thus increased fragility leading to higher propensity of the premature fetus for germinal matrix hemorrhage and other intracerebral bleeds; however, further investigation is needed to examine its role during fetal life. Similarly, extracellular superoxide dismutase (SOD) expression was significantly down-regualted during early life, compared to adult. Vascular tissue express 3 distinct isoforms of SOD: cytosolic or copper-zinc SOD (CuZn-SOD; SOD1), manganese SOD (Mn-SOD) localized in mitochondria (mitochondrial SOD or SOD2), and an extracellular form of CuZn-SOD (EC-SOD; SOD3) (20). Because there are no selective pharmacological inhibitors of individual SOD isoforms, the functional importance of the different SODs has been difficult to define. However, the present finding of significantly down-regulated expression of specifically EC-SOD3 suggest an distinct role in early life. SOD plays a crucial role in conversion of superoxide anion ( O2·−) to H2O2; which is further converted to H2O by the actions of glutathione peroxidases and peroxiredoxins. Of importance, in the present study, we observed that along with reduced expression of SOD3, there was a significantly increased expression of both glutathione peroxidase (GPX8) and peroxiredoxin4 (PRDX4). Thus, the system is geared towards reduced production and rapid clearance of H2O2. At present, the clear rational of this is not known.
Cell proliferation, growth, and assembly pathway genes expression is up-regulated during early life
To no surprise, many genes involved in cell cycle regulation, DNA replication, chromosome assembly, and other components of CA cell replication, growth, and assembly demonstrated increased expression. However of importance, the fold-changes of these genes were to a significantly greater in the premature fetus and newborn than in near-term fetus. This emphasizes the complexity of the changes, which occur at the several developmental ages. The present study also demonstrates that aurora kinase A and B, several cyclins, centromere proteins and replication factors are significantly up-regulated during early life. Over expression of Aurora A and Aurora B can lead to genetic instability (gain or loss of whole chromosomes) by the over phosphorylation of normal cell cycle targets and the aberrant phosphorylation of cytoplasmic targets (21). The resultant chromosomal instability is a common feature of many cancer types (22). A significant over-expression of these kinases during fetal life, however, contradicts these findings, and suggests that the precise role during early life requires further investigation.
Moreover, ubiquitin conjugating enzyme e2c (UBE2C) was up-regulated almost 260-fold in the premature fetus and its expression fell to ~ 66-fold near-term.. Nonetheless, this indicates high expression of this gene during early life. Importantly, excessive UBE2C is known to disrupt normal chromosome segregation, or even lead to mis-separation of the chromosomes (23) and may lead to malignancies (24). The significance of such high levels in the early life is unknown, however.
MAPK-ERK signaling pathway gene expression is down-regulated in early life
The mitogen activated protein kinase (MAPK) pathway is a major signaling-cascade involved in the cellular growth and development. In the present studies, several components of the MAPK pathway including MAPK3, MAP2K2, MAP kinase interacting serine/threonine kinase 1 and 2 were expressed several folds lower during early life. Of importance, we also observed a significant reduction in the expression of Protein Kinase C – Delta (PRKCD) in fetus and newborn, compared to adult. Recent studies demonstrate that upstream to the MAPK pathway, PRKCD is necessary for the activation of MAPK3 (25). Thus, the findings suggest that several components of MAPK-ERK pathway are suppressed during early life. Importantly, evidence supports the idea that down-regulation of this pathway is advantageous for organism survival and well being. For instance, MAPK3 down-regulation has been demonstrated to be beneficial for striatum-dependent long-term memory (26), reduced adiposity and protection from high-fat diet induced obesity and insulin resistance (27). Similar results of MAPK-mediated negative regulation of self-renewal cell divisions have been demonstrated in developing plant cells (28).
Based on the present study and the above-mentioned reports, reduced expression and down-regulation of MAPK3 would appear to play a critical role in development. In previous reports, we have demonstrated that MAPK plays a significantly reduced role in cerebral arterial contractility during fetal life, compared to that in adult (29). Nonetheless, the MAPK cascade involvement in negative regulation of vascular growth and development necessitates further investigation.
Actin cytoskeleton pathway genes expression is down-regulated in early life
In CA several important members of the actin cytoskeleton canonical pathways were demonstrated reduced expression during early life, compared to the adult. Of note filamin A (FLNA), expression was down-regulated to a significantly greater extent in premature than that in near-term fetal arteries. The reduced expression of FLNA was significantly more pronounced on real-time PCR analysis, compared to that of microarray analysis. Several explanations could account for this. For instance, microarray examination is based on the ratio of the densitometric analysis of the signal, whereas in PCR the florescence signal is exponentially weighted to the power of 2 to correct for the doubling with each cycle. This makes the PCR technique much more sensitive compared to the microarray analysis. Nonetheless, with both techniques, the finding that there is a significantly greater reduction of FLNA in the premature vessels remains the same. Studies have demonstrated that filamin exists in three isoforms, FLNA, filamin B (FLNB), and filamin C (FLNC) (30). Of critical importance, FLNC has been shown to have a restricted expression in skeletal and cardiac muscle (30) and FLNA and FLNB play a crucial role in corticogenesis and brain development (31). However, the present study shows significant greater reduction of FLNA expression in premature CA, compared to near-term fetus. A clear rationale of such a finding currently is unknown. Nonetheless, the present study suggests an important role of FLNA in vascular development. Our results demonstrate further that Formin Binding Protein 1 (FBP1) was reduced significantly during early life. FBP1 has been implicated in smooth muscle phenotype switching to a contractile type, and this agrees with findings of the present study (32). However, the actin cytoskeleton signaling cascade has been implicated in cell motility, surface remodeling, cell shape changes during mitosis, muscle contraction, separation of daughter cells by the contractile ring during cytokinesis, cell-cell and cell-substrate interactions together with adhesion molecules, trans-membrane signaling, endocytosis, and secretion (33). Down-regulation of this pathway in carotid arteries may suggest that it plays a critical role in the above-mentioned aspects of smooth muscle contractile and/or development pathway.
Integrin signaling pathway gene expression is down-regulated during early life
Another important signaling pathway altered with development was that of integrin signaling (Table 8). The present study demonstrates that major components of the integrin-signaling cascade expression was down-regulated in both immature and near-term fetus, as well as newborn CA. Integrins have been implicated in several fundamental cellular functions such as cellular movement, stabilization, and adhesion (34), as well as internal cellular cytoskeleton organization (35). Evidence also supports the idea that integrins through janus kinase (JAK)/signal transducer and activator of transcription (STAT) and MAPK pathways mediate intracellular signaling (36). Not only integrins, but also their down-stream mediators such as the MAPK pathway expression was down-regulated. In contrast, caveolin, a scaffolding protein which is a part of the integrin signaling cascade, was expressed to a significantly greater extent in premature fetus and newborn, whereas its expression was reduced in near-term fetus. Of note, caveolin links integrin subunits to the tyrosine kinase FYN, an initiating step in coupling integrins to the Ras-ERK pathway and promoting cell cycle progression (37); moreover, it is a negative regulator of the Ras-MAPK cascade (38). As noted, the MAPK pathway expression is down-regulated in early fetal life, and caveolin, a negative regulator of this pathway expression is up-regulated. This suggests an active down-regulation of the MAPK cascade and further indicates that MAPK inhibition plays a critical role during early life.
Perspective.
Overall, results of the present study demonstrate significant alterations of gene expression with maturation of the cranial vasculature. For the first time, we demonstrate that, compared to more active gene expression of several signaling pathways in the premature fetus and newborn organism, in the near-term fetus differential gene expression is attenuated significantly. These findings agree with the concept of molecular mechanisms acting in an integrated manner to regulate both phenotypic and mechanical plasticity in vascular smooth muscle cells (39) and may be an important step for the prepration of the fetus for birth. Moreover, the present study raises a number of questions regarding the regulation of changes in vascular gene expression and their biological significance in the developing premature fetus, near-term fetus, and newborn. The present study also provides a basis for future studies to explore the importance of changes in the major signal transduction pathways during early vascular development and their function. Perhaps most importantly, this study suggests avenues in which to target the developing cerebral vasculature for gene therapy to ammeliorate the pathophysiologic disruptions that may occur during early life (40).
Acknowledgments
Financial Support: The studies were supported by USPHS Grant HD-03807 to LDL
We would like to acknowledge Nina Chu, Dipali Goyal, Nathanael Matei, and Giovanni A. Longo for their expert technical support. We would also like to acknowledge staff of GenUs Biosystems for help with the microarray experiments and analysis.
Reference Cited
- 1.Donnan GA, Fisher M, Macleod M, Davis SM. Stroke. Lancet. 2008;371:1612–1623. doi: 10.1016/S0140-6736(08)60694-7. [DOI] [PubMed] [Google Scholar]
- 2.Sheth RD. Trends in incidence and severity of intraventricular hemorrhage. J Child Neurol. 1998;13:261–264. doi: 10.1177/088307389801300604. [DOI] [PubMed] [Google Scholar]
- 3.Nelson KB. Can we prevent cerebral palsy? N Eng J Med. 2003;349:1765–1769. doi: 10.1056/NEJMsb035364. [DOI] [PubMed] [Google Scholar]
- 4.Donegan JH, Traystman RJ, Koehler RC, Jones MD, Rogers MC. Cerebrovascular hypoxic and autoregulatory responses during reduced brain metabolism. Am J Physiol. 1985;249:H421–9. doi: 10.1152/ajpheart.1985.249.2.H421. [DOI] [PubMed] [Google Scholar]
- 5.Heistad DD, Marcus ML, Abboud FM. Role of large arteries in regulation of cerebral blood flow in dogs. J Clin Invest. 1978;62:761–768. doi: 10.1172/JCI109187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dieckhoff D, Kanzow E. On the location of the flow resistance in the cerebral circulation. Pflugers Archiv: Eu J Physiol. 1969;310:75–85. doi: 10.1007/BF00586876. [DOI] [PubMed] [Google Scholar]
- 7.Kontos HA, Wei EP, Navari RM, et al. Responses of cerebral arteries and arterioles to acute hypotension and hypertension. Am J Physiol. 1978;234:H371–83. doi: 10.1152/ajpheart.1978.234.4.H371. [DOI] [PubMed] [Google Scholar]
- 8.Bassan H. Intracranial hemorrhage in the preterm infant: understanding it, preventing it. Clin Perinatol. 2009;36:737–62. v. doi: 10.1016/j.clp.2009.07.014. [DOI] [PubMed] [Google Scholar]
- 9.Ward Platt M, Deshpande S. Metabolic adaptation at birth. Semin Fetal Neonatal Med. 2005;10:341–350. doi: 10.1016/j.siny.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Wagerle LC, Moliken W, Russo P. Nitric oxide and beta-adrenergic mechanisms modify contractile responses to norepinephrine in ovine fetal and newborn cerebral arteries. Pediatr Res. 1995;38:237–242. doi: 10.1203/00006450-199508000-00017. [DOI] [PubMed] [Google Scholar]
- 11.Goyal R, Mittal A, Chu N, et al. Maturation and long-term hypoxia-induced acclimatization responses in PKC-mediated signaling pathways in ovine cerebral arterial contractility. Am J Physiol Regul Integr Comp Physiol. 2010;299:R1377–86. doi: 10.1152/ajpregu.00344.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goyal R, Creel KD, Chavis E, et al. Maturation of intracellular calcium homeostasis in sheep pulmonary arterial smooth muscle cells. Am J Physiol Lung Cell Mol Physiol. 2008;295:L905–14. doi: 10.1152/ajplung.00053.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goyal R, Mittal A, Chu N, Zhang L, Longo LD. alpha(1)-Adrenergic receptor subtype function in fetal and adult cerebral arteries. Am J Physiol Heart Circ Physiol. 2010;298:H1797–806. doi: 10.1152/ajpheart.00112.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goyal R, Yellon SM, Longo LD, Mata-Greenwood E. Placental gene expression in a rat “model” of placental insufficiency. Placenta. 2010;31:568–575. doi: 10.1016/j.placenta.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 15.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szymonowicz W, Walker AM, Yu VY, et al. Regional cerebral blood flow after hemorrhagic hypotension in the preterm, near-term, and newborn lamb. Pediatr Res. 1990;28:361–366. doi: 10.1203/00006450-199010000-00012. [DOI] [PubMed] [Google Scholar]
- 17.LeClair R, Lindner V. The role of collagen triple helix repeat containing 1 in injured arteries, collagen expression, and transforming growth factor beta signaling. Trends Cardiovasc Med. 2007;17:202–205. doi: 10.1016/j.tcm.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Tang L, Dai DL, Su M, et al. Aberrant expression of collagen triple helix repeat containing 1 in human solid cancers. Clin Cancer Res. 2006;12:3716–3722. doi: 10.1158/1078-0432.CCR-06-0030. [DOI] [PubMed] [Google Scholar]
- 19.Goyal R, Henderson DA, Chu N, Longo LD. Ovine middle cerebral artery characterization and quantification of ultrastructure and other features: changes with development. Am J Physiol Regul Integr Comp Physiol. 2012 doi: 10.1152/ajpregu.00519.2011. Epub; ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faraci FM, Didion SP. Vascular protection: superoxide dismutase isoforms in the vessel wall. Arterio Thromb Vas Biol. 2004;24:1367–1373. doi: 10.1161/01.ATV.0000133604.20182.cf. [DOI] [PubMed] [Google Scholar]
- 21.Carvajal RD, Tse A, Schwartz GK. Aurora kinases: new targets for cancer therapy. Clin Cancer Res. 2006;12:6869–6875. doi: 10.1158/1078-0432.CCR-06-1405. [DOI] [PubMed] [Google Scholar]
- 22.McClellan WJ, Dai Y, Abad-Zapatero C, et al. Discovery of potent and selective thienopyrimidine inhibitors of Aurora kinases. Bioorg Med Chem Lett. 2011;21:5620–5624. doi: 10.1016/j.bmcl.2011.06.041. [DOI] [PubMed] [Google Scholar]
- 23.Skibbens RV, Hieter P. Kinetochores and the checkpoint mechanism that monitors for defects in the chromosome segregation machinery. Annu Rev Genet. 1998;32:307–337. doi: 10.1146/annurev.genet.32.1.307. [DOI] [PubMed] [Google Scholar]
- 24.van Ree JH, Jeganathan KB, Malureanu L, van Deursen JM. Overexpression of the E2 ubiquitin-conjugating enzyme UbcH10 causes chromosome missegregation and tumor formation. J Cell Biol. 2010;188:83–100. doi: 10.1083/jcb.200906147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuriyama M, Taniguchi T, Shirai Y, et al. Activation and translocation of PKCdelta is necessary for VEGF-induced ERK activation through KDR in HEK293T cells. Biochem Biophys Res Commun. 2004;325:843–851. doi: 10.1016/j.bbrc.2004.10.102. [DOI] [PubMed] [Google Scholar]
- 26.Mazzucchelli C, Vantaggiato C, Ciamei A, et al. Knockout of ERK1 MAP kinase enhances synaptic plasticity in the striatum and facilitates striatal-mediated learning and memory. Neuron. 2002;34:807–820. doi: 10.1016/s0896-6273(02)00716-x. [DOI] [PubMed] [Google Scholar]
- 27.Bost F, Aouadi M, Caron L, et al. The extracellular signal-regulated kinase isoform ERK1 is specifically required for in vitro and in vivo adipogenesis. Diabetes. 2005;54:402–411. doi: 10.2337/diabetes.54.2.402. [DOI] [PubMed] [Google Scholar]
- 28.Lampard GR, Lukowitz W, Ellis BE, Bergmann DC. Novel and expanded roles for MAPK signaling in Arabidopsis stomatal cell fate revealed by cell type-specific manipulations. Plant Cell. 2009;21:3506–3517. doi: 10.1105/tpc.109.070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goyal R, Mittal A, Chu N, et al. Maturation and the role of PKC-mediated contractility in ovine cerebral arteries. Am J Physiol Heart Circ Physiol. 2009;297:H2242–52. doi: 10.1152/ajpheart.00681.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng Y, Walsh CA. The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat Cell Biol. 2004;6:1034–1038. doi: 10.1038/ncb1104-1034. [DOI] [PubMed] [Google Scholar]
- 31.Sheen VL, Feng Y, Graham D, et al. Filamin A and Filamin B are co-expressed within neurons during periods of neuronal migration and can physically interact. Hum Mol Genet. 2002;11:2845–2854. doi: 10.1093/hmg/11.23.2845. [DOI] [PubMed] [Google Scholar]
- 32.Staus DP, Blaker AL, Medlin MD, Taylor JM, Mack CP. Formin homology domain-containing protein 1 regulates smooth muscle cell phenotype. Arterio Thrombo Vas Biol. 2011;31:360–367. doi: 10.1161/ATVBAHA.110.212993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidt A, Hall MN. Signaling to the actin cytoskeleton. Annu Rev Cell Dev Biol. 1998;14:305–338. doi: 10.1146/annurev.cellbio.14.1.305. [DOI] [PubMed] [Google Scholar]
- 34.Campbell ID, Humphries MJ. Integrin structure, activation, and interactions. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a004994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suen PW, Ilic D, Caveggion E, et al. Impaired integrin-mediated signal transduction, altered cytoskeletal structure and reduced motility in Hck/Fgr deficient macrophages. J Cell Sci. 1999;112 ( Pt 22):4067–4078. doi: 10.1242/jcs.112.22.4067. [DOI] [PubMed] [Google Scholar]
- 36.Cabodi S, Di Stefano P, Leal MDPC, et al. Integrins and signal transduction. Adv Exp Med Biol. 2010;674:43–54. doi: 10.1007/978-1-4419-6066-5_5. [DOI] [PubMed] [Google Scholar]
- 37.Wary KK, Mariotti A, Zurzolo C, Giancotti FG. A requirement for caveolin-1 and associated kinase Fyn in integrin signaling and anchorage-dependent cell growth. Cell. 1998;94:625–634. doi: 10.1016/s0092-8674(00)81604-9. [DOI] [PubMed] [Google Scholar]
- 38.Nonami A, Taketomi T, Kimura A, et al. The Sprouty-related protein, Spred-1, localizes in a lipid raft/caveola and inhibits ERK activation in collaboration with caveolin-1. Genes Cells. 2005;10:887–895. doi: 10.1111/j.1365-2443.2005.00886.x. [DOI] [PubMed] [Google Scholar]
- 39.Halayko AJ, Solway J. Molecular mechanisms of phenotypic plasticity in smooth muscle cells. J Appl Physiol. 2001;90:358–368. doi: 10.1152/jappl.2001.90.1.358. [DOI] [PubMed] [Google Scholar]
- 40.Watanabe Y, Heistad DD. Targeting cerebral arteries for gene therapy. Exp Physiol. 2005;90:327–331. doi: 10.1113/expphysiol.2004.028498. [DOI] [PubMed] [Google Scholar]