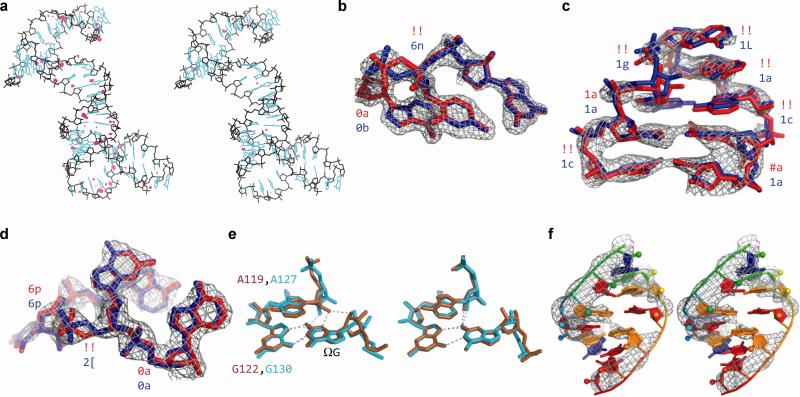
Figure 1.
Examples of geometric improvements by ERRASER-PHENIX. (a) Clash reduction in 3R4F. Red dots: unfavorable clashes. Left: PDB. Right: ERRASER-PHENIX. (b, c, d) Backbone conformation improvement on (b) nucleotides 62-64, chain A of 1U8D, (c) nucleotides 27-34, chain Q of 2OIU and (d) nucleotides 33-36, chain A of 2YGH. Rotamer assignments are shown at each suite. ‘!!’ stands for outlier suites. Red: PDB. Blue: ERRASER-PHENIX. (e) Functionally relevant pucker correction on group I ribozyme models. Brown: 1Y0Q. Cyan: 3BO3. Left: PDB. Right: ERRASER-PHENIX. (f) Base-pair geometry improvement on nucleotides 1-6 and 66-71, chain A of 3P49. Left: PDB. Right: ERRASER-PHENIX.