FIGURE 1.
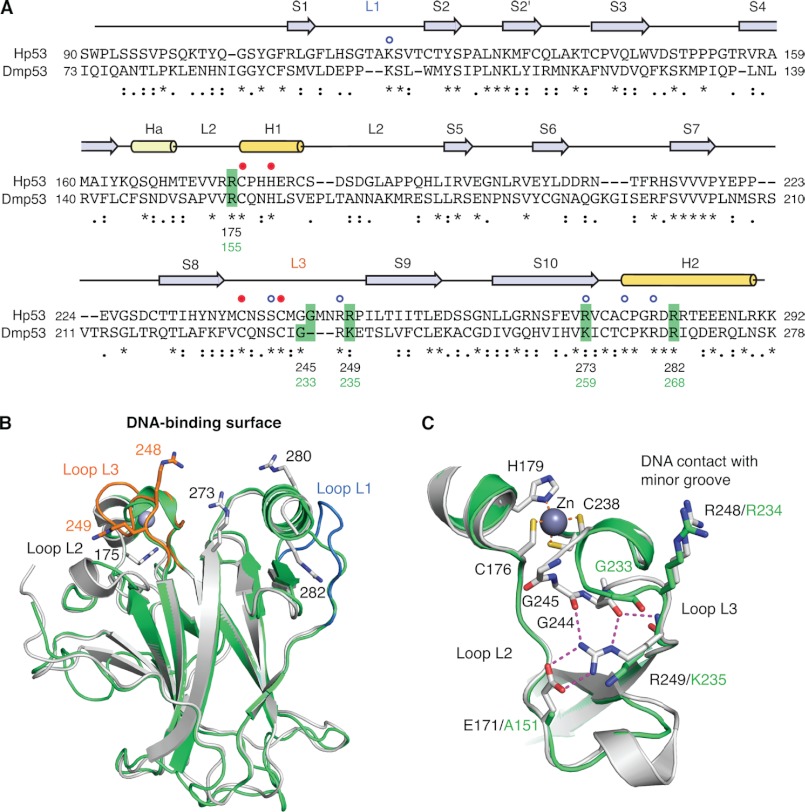
Comparison of the DNA-binding domains of human and Drosophila p53. A, structure-based sequence alignment of Hp53 and Dmp53 DBD. Sequence conservation is indicated below the alignment: * denotes conserved residues, : denotes similar residues, and · denotes somewhat similar residues as classified by CLUSTALW2 (44). Secondary structure elements of Hp53 (S, β-strand; H1/2, α-helix; Ha, 310-helix; L, Loop) are shown above the alignment based on Protein Data Bank (PDB) entry 2XWR (45). The blue open circles denote Hp53 DNA contact residues, and the closed red circles indicate the residues coordinating the zinc ion. Dmp53 residues that were mutated in this study and the corresponding cancer hot spot sites in Hp53 are highlighted in green. B, superposition of the crystal structure of Hp53 DBD (gray ribbon diagram; PDB entry 2XWR) (45) and a homology model of the Dmp53 DBD (green ribbon diagram). Loops L1 and L3 of Hp53 are highlighted in blue and orange, respectively. The side chains of a number of structurally and functionally important arginine residues of Hp53 are shown as stick models. C, superposition of the L2/L3 region in Hp53 (gray) and Dmp53 (green) showing large differences in the L3 loop conformation. The extensive polar interaction network of Arg-249 in Hp53 that stabilizes the hairpin conformation of L3 is shown by broken lines.