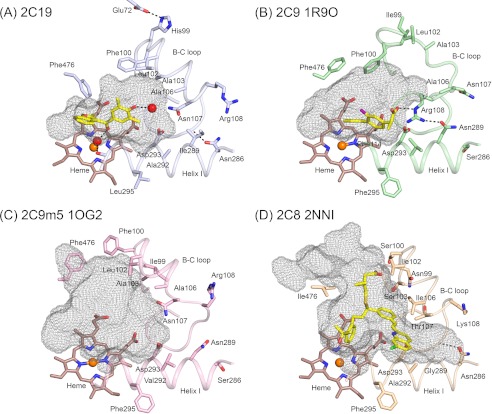
FIGURE 4.
Conformational differences between helix B-C loops significantly alter the active site cavities of the structure of the P450 2C19 0XV complex (A), the structure of the flurbiprofen complex of 2C9, PDB code 1R9O (B), the apo 2C9m7 structure, PDB code 1OG2 (C), and the 2C8 structure, PDB code 2NNI, of the montelukast complex (D). Substrate-binding cavities were calculated with Voidoo and rendered as mesh surfaces. A water molecule represented as a red sphere is hydrogen-bonded (dashed line) to the 0XV hydroxyl moiety, and a second water molecule is bound to the heme iron of chain A, where it forms hydrogen bonds (dashed lines) with the carbonyl of 0XV and the carbonyl of Ala-297 of 2C19. Additional hydrogen bonds discussed in the text are indicated by dashed lines. Amino acid residues that contribute to differential substrate selectivity and/or that directly shape the distal portions of the cavity are displayed as sticks. Color coding is the same as described for Figs. 2 and 3 with the additional use of yellow and purple to color the carbon and fluorine atoms, respectively, of flurbiprofen in the 2C9 1R9O structure.