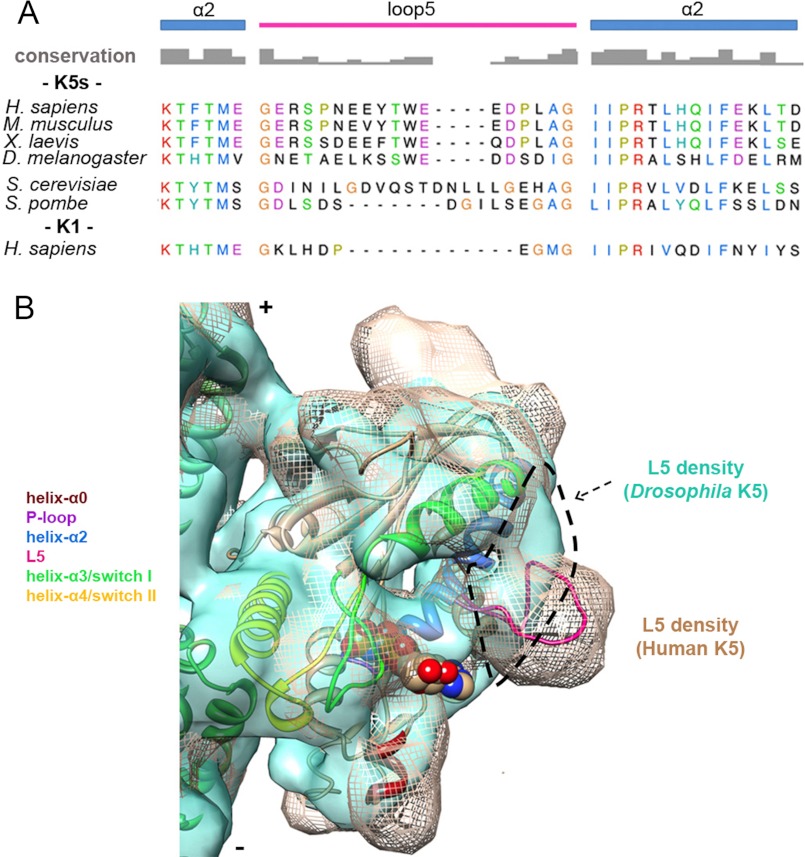
FIGURE 6.
Comparison of K5s L5 sequence and conformation. A, alignment of helix-α2/L5 sequences from K5s (Homo sapiens P52732, 111–149; Mus musculus Q6P9P6, 110–148; Xenopus laevis P28025, 104–142; D. melanogaster P46863, 109–147; Saccharomyces cerevisiae P28742, 137–189; Schizosaccharomyces pombe P24339, 155–200) and K1 (H. sapiens P33176, 91–121) generated using T-coffee (63). Secondary structure elements and amino acid conservation are indicated. Among these sequences, identical or similar residues are shown with the same color, whereas non-conserved residues are in black. B, cryo-EM reconstruction of MT-bound Drosophila K5 MD (KLP61F)-AMPPNP (light green surface, 0.5 σ, Electron Microscopy Data Bank entry 1604 (29)) superposed on the human K5 MD-AMPPNP structure (mesh, 1.1 σ). Our pseudoatomic AMPPNP MD model is fitted in the mesh map. The dashed outline delineates loop 5 density in KLP61F reconstruction showing the very different conformations of L5 in these K5s.