Background: Studies on the diversity of carbohydrate-binding proteins (lectins) are important in glycobiology.
Results: A lectin having a novel primary structure was isolated from a mussel and found to have a globotriose-dependent cytotoxicity on Burkitt lymphoma cells.
Conclusion: A new primary structure quite distinct from known lectin is described.
Significance: Discovery of similar lectin structures from vertebrates will lead to progress in medical sciences.
Keywords: Carbohydrate-binding Protein, Cell Death, Galactose, Glycolipids, Glycomics, Lectin, Protein Chemistry, Protein Purification, Globotriaosylceramide, Mussel
Abstract
A novel lectin structure was found for a 17-kDa α-d-galactose-binding lectin (termed “MytiLec”) isolated from the Mediterranean mussel, Mytilus galloprovincialis. The complete primary structure of the lectin was determined by Edman degradation and mass spectrometric analysis. MytiLec was found to consist of 149 amino acids with a total molecular mass of 16,812.59 Da by Fourier transform-ion cyclotron resonance mass spectrometry, in good agreement with the calculated value of 16,823.22 Da. MytiLec had an N terminus of acetylthreonine and a primary structure that was highly novel in comparison with those of all known lectins in the structure database. The polypeptide structure consisted of three tandem-repeat domains of ∼50 amino acids each having 45–52% homology with each other. Frontal affinity chromatography technology indicated that MytiLec bound specifically to globotriose (Gb3; Galα1–4Galβ1–4Glc), the epitope of globotriaosylceramide. MytiLec showed a dose-dependent cytotoxic effect on human Burkitt lymphoma Raji cells (which have high surface expression of Gb3) but had no such effect on erythroleukemia K562 cells (which do not express Gb3). The cytotoxic effect of MytiLec was specifically blocked by the co-presence of an α-galactoside. MytiLec treatment of Raji cells caused increased binding of anti-annexin V antibody and incorporation of propidium iodide, which are indicators of cell membrane inversion and perforation. MytiLec is the first reported lectin having a primary structure with the highly novel triple tandem-repeat domain and showing transduction of apoptotic signaling against Burkitt lymphoma cells by interaction with a glycosphingolipid-enriched microdomain containing Gb3.
Introduction
Mollusks are important aquatic and scientific resources. The Mediterranean mussel (Mytilus galloprovincialis; family Mytilidae) is an invasive species that originated in the Mediterranean and has been introduced to intertidal and near-shore habitats in many parts of the world, including the coasts of Japan. These marine bivalves are filter feeders that filter large amounts of debris, often including pathogenic microorganisms or heavy metals, and have evolved tolerance and defense mechanisms that help them adapt to diverse environments. Recent genomic research on bivalves has led to the establishment of expressed sequence tag libraries (1–3). One of these libraries has been useful for the identification of key genes that regulate pearl formation in pearl oysters (1). Another expressed sequence tag library, MytiBase, developed from studies of M. galloprovincialis, provides a valuable bioinformatics tool for investigating mechanisms involved in development, differentiation, and defense in this species. The MytiBase library has been useful in the elucidation of novel genes expressed in hemocytes and related to innate immunity in studies of Vibrio bacterial infection (3).
Lectins are glycan-binding proteins that function in the recognition of a wide variety of glycan structures and can be used to decipher the glyco-codes that determine the composition of oligosaccharides in glycoconjugates (glycosphingolipids, glycoproteins, and proteoglycans). Lectins have been isolated from almost all animal and plant phyla. Certain lectin-coding genes in bivalves have been found to be up- or down-regulated in association with infection by pathogenic microorganisms, indicating that the lectins are able to respond to external/environmental stimuli (3, 4). Several lectins with characteristic structures have been described in bivalves, including C-type lectins and galectins (5, 6), fibrinogen-type (7), C1q-type (8), and F-type lectins (fucolectin) (9). A Gal/GalNAc-binding lectin (information on primary structure unavailable) was reported in Crenomytilus, a genus related to Mytilus (10). A lectin domain of the sea urchin egg lectin (SUEL)3-type that was originally reported in a deuterostome (sea urchin) (11) was later identified in the bivalve Pteria penguin (12). These findings suggest that bivalves, including mytilids, are interesting subjects for studies of characteristic lectins and glycan-dependent phenomena. Glycobiological investigation of these animal will help elucidate their biochemical and physiological mechanisms.
A variety of methodologies using advanced equipment has been developed to elucidate the glycan binding specificities of lectins, to identify the specific oligosaccharides involved, and to better understand the molecular interactions between lectins and glycans (13–16, 19, 20). The precise glycan binding specificities of lectins have been identified using sophisticated glycome procedures such as glycan microarrays (13, 14) and frontal affinity chromatography technology (FACT) (15–20). In FACT analysis, a column-immobilized lectin is connected to an HPLC pump, and fluorescence detector and pyridylamino (PA)-labeled oligosaccharides are injected onto the column. The affinity of glycan binding to the lectin is assessed based on the delaying elution volume of the oligosaccharides. Studies using FACT analysis have revealed that the diversity of glycan-binding profiles for d-Gal-binding lectins in aquatic animals is much greater than was previously suspected (12, 21–25).
In this study, we purified an α-Gal-binding lectin from the mantle of M. galloprovincialis. Protein chemical analysis and glycome procedures revealed that the primary structure of the lectin is extremely novel and has no similarity to previously described structures. The unique glycan-binding profile of the lectin involves a specific affinity with a neutral glycan in the glycosphingolipid Gb3 (Galα1–4Galβ1–4Glc). The lectin displayed Gb3-dependent cytotoxicity against Burkitt lymphoma cells that express the glycan.
EXPERIMENTAL PROCEDURES
Animal, Cells, and Chemicals
Mussels (M. galloprovincialis) were obtained from Hirakata Bay and Blue Carbon experimental raft (the city of Yokohama) in Tokyo Bay, Yokohama, Kanagawa Prefecture, Japan. The shells were removed, and the mantles were stored at −80 °C. Human Burkitt lymphoma Raji cells and erythroleukemia K562 cells were from the Cell Resource Center for Biomedical Research, Institute of Development, Aging, and Cancer, Tohoku University (Sendai, Japan), and the Japanese Cancer Research Resources Bank (Tokyo, Japan), respectively. Lysyl endopeptidase, phenylmethylsulfonyl fluoride, lactose, melibiose, sucrose, d-galactose, d-glucose, d-mannose, d-fucose, l-fucose, d-talose, d-gulose, GalNAc, and GlcNAc, each of the highest purity grade, were from Wako Pure Chemical Co., Tokyo, Japan. Methyl α-d-galactopyranoside, methyl β-d-galactopyranoside, methyl α-N-acetyl d-galactosaminide, and methyl β-N-acetyl d-galactosaminide were from Pfanstiehl Laboratories, Waukegan, IL. Fetuin, asialofetuin, bovine submaxillary mucin, asialo-bovine submaxillary mucin, and standard protein markers for gel permeation chromatography (GPC) were from Sigma. Melibiosyl-agarose gel and protease inhibitor mixture were from Cosmo Bio Co., Tokyo, Japan. Superdex 75, Sephadex G-75, N-hydroxysuccinimide-activated Sepharose Fast Flow, sensor chip CM5, and ligand coupling kit were from GE Healthcare. Neoglycoprotein globotriaosyl (Gb3; Galα1–4Galβ1–4Glc)-human serum albumin (Gb3-HSA) was from Carbosynth Ltd., Berkshire, UK. A standard protein marker mixture for SDS-PAGE, Pfu N-acetyl deblocking aminopeptidase, endoproteinase Asp-N, and 15 PA-oligosaccharides were from Takara Bio Inc., Kyoto, Japan. Cell counting kit-8, including 2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodiumsalt (WST-8), and HEPES were from Dojindo Co., Kumamoto, Japan. RPMI 1640 medium was from Nissui Pharmaceutical Co., Tokyo, Japan. Fetal calf serum was from Invitrogen. Penicillin/streptomycin was from Roche Diagnostics. Trypan blue solution was from Nacalai Tesque, Inc., Kyoto, Japan. MEBCYTO apoptosis kit was from Medical & Biological Laboratories Co., Nagoya, Japan. FACSCalibur was from BD Biosciences. GLOMAX MultiDetection System was from Promega, Madison, WI.
Lectin Purification
Mantles were homogenized with 10 volumes (w/v) of 150 mm NaCl containing 10 mm Tris-HCl, pH 7.5 (TBS), 10 mm EDTA, 5 mm benzamidine, and 2 mm PMSF. The supernatant (Sup 1) was collected by centrifugation at 27,500 × g for 1 h at 4 °C. The precipitate was homogenized with 10 volumes (w/v) of 100 mm d-Gal containing TBS, and the supernatant (Sup 2) was collected as above. Sup 2 was dialyzed extensively against TBS. Sup 1 and Sup 2 were both applied to a melibiosyl-agarose column (5.0 ml), and the column was washed with TBS until the absorbance of the effluent at 280 nm reached the base-line level. The lectin was eluted with TBS containing 200 mm melibiose.
Hemagglutination Assay and Sugar Binding Specificity
Hemagglutination assay was performed in 96-well V-shape plates as described previously (26). Twenty μl of a 2-fold dilution of purified lectin in TBS was mixed with 20 μl of a 1% suspension (with TBS; v/v) of trypsinized and glutaraldehyde-fixed rabbit erythrocytes, 20 μl of TBS, and 20 μl of TBS with 1% Triton X-100. The plate was incubated at room temperature for 1 h, and the formation of a sheet (agglutination-positive) or dot (agglutination-negative) was observed and scored as the lectin titer. For analysis of sugar binding specificity, 20 μl of each sugar solution (prepared at 200 mm) was serially diluted with TBS, mixed with 20 μl each of the lectin solution (previously adjusted to titer 16), trypsinized, and glutaraldehyde-fixed with rabbit erythrocytes and TBS containing 1% Triton X-100. The plate was incubated at room temperature for 1 h, and the minimal inhibitory sugar concentration was determined.
Protein Determination
Protein was quantified using a protein assay kit (Pierce) based on the principle of bicinchoninic acid for colorimetric detection (27, 28), using ovalbumin as a standard. SDS-PAGE (29) was performed in 15% (w/v) acrylamide gel under reducing or nonreducing conditions. The gel was stained by either Coomassie Brilliant Blue or R-250.
Gel Permeation Chromatography
The purified lectin was dissolved and subjected to GPC on a Superdex 75 column (1.0 × 32 cm) connected to an HPLC system consisting of a PU-2089 intelligent pump and a UV-2027 UV-visible detector (Jasco Co., Tokyo, Japan). Standard molecular mass marker proteins and purified lectin were separated at a flow rate of 0.5 ml/min in 100 mm melibiose containing TBS. Proteins were detected at an absorbance of 280 nm using the UV detector.
Enzymatic Digestion and Chemical Cleavage
Reversed-phase HPLC- purified lectin (2–5 nmol) was digested in 100 mm Tris-HCl buffer containing 2 m urea, pH 9.0, with lysyl endopeptidase (30) at an enzyme-to-substrate molar ratio of 1:300 or in 50 mm Tris-HCl buffer containing 2 m urea, pH 8.5, with endoproteinase Asp-N at an enzyme-to-substrate molar ratio of 1:20, at 37 °C for 18 h. The lectin (2 nmol) was chemically cleaved at the methionyl bonds with 2% CNBr in 70% (v/v) formic acid at 25 °C for 18 h in the dark by the method of Gross (31). The peptide with the amino-terminal blocking group was treated with the deblocking aminopeptidase N-acetyl deblocking aminopeptidase in 50 mm N-ethylmorpholine/acetic acid buffer containing 0.1 mm CoCl2, pH 7.5, at 37 °C for 20 h, according to the manufacturer's instructions (32).
N-O-Acyl Migration of the Blocked Amino-terminal Peptide
Twenty pmol of the blocked amino-terminal peptide, K1 (residue 1–6), was dried in a polypropylene tube and incubated with 10 ml of 37% HCl for 20 h in an N2 atmosphere at room temperature (33). The dried reaction mixture was subjected to automated Edman degradation.
Separation of Peptides
Peptides generated by enzymatic digestion or by CNBr cleavage as above were separated by RP-HPLC using a Hewlett Packard model 109M liquid chromatograph on an Aquapore RP-300 (2.1 × 30 mm, Applied Biosystems, Inc., Carlsbad, CA), a Mightysil RP-18 (2.0 × 50 mm, Kanto Chemical Co., Tokyo, Japan), and a Superspher Select B column (2.0 × 119 mm, Merck). Each column was equilibrated with solvent A (0.09% TFA), and peptides were eluted at a flow rate of 200 μl/min using a linear gradient of 0–80% solvent B (acetonitrile/water/TFA, 80:20:0.075 (v/v)) at room temperature. Peptides generated by CNBr cleavage were also separated by GPC-HPLC on tandem SynChropak GPC-Peptide columns (4.6 × 250 mm, SynChrom Inc., Linden, IN) connected to a Hewlett Packard 1040 M detection system in 0.1 m sodium phosphate containing 6 m guanidine hydrochloride. The eluent was monitored with a diode array detector, and the separated fractions were purified by RP-HPLC as described above.
Amino Acid Composition and Sequence Analysis
The purified whole lectin or its peptide was hydrolyzed with 6 n HCl containing 0.1% (w/v) phenol at 110 °C for 20 h by the vapor phase method or in 4 n methanesulfonic acid containing 0.2% tryptamine (3-(2-aminoethyl) indole) for 20 h at 110 °C (34). The amino acid composition was analyzed using a model L8500 amino acid analyzer (Hitachi). Automated Edman degradation was performed with a gas phase protein sequencer (Applied Biosystems model 477A) (35). On-line sequence homology searches (www.ncbi.nlm.nih.gov) were performed using either the blastp or tblastn program with the nonredundant databases of the National Center for Biotechnology Information (NCBI), Bethesda (36, 37).
Mass Spectrometry
Matrix-associated laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) analysis of the whole protein and of each peptide was performed using REFLEX equipment (Bruker-Franzen Analytik, Bremen, Germany). An aliquot (0.5 μl) of each HPLC fraction was spotted onto a target and added with 0.5 μl of 2-mercaptobenzothiazole (38) dissolved in ethanol/water (1:1, v/v) as matrix. The mixed samples were dried at room temperature for 5 min, and each spot was desorbed with 80 laser shots for positive-mode analysis. To determine the precise molecular mass of the whole protein and the selected peptides, samples were diluted with 4 volumes of 50% methanol containing 3% acetic acid and subjected to electron spray ionization Fourier transform ion cyclotron resonance mass spectrometry (ESI/FT-ICR MS) (39). Mass spectra were acquired with a BioAPEX 7.0 (Bruker Instruments, Germany) equipped with an external electrospray ionization source (Analytica of Branford, Inc., Branford, CT) and a syringe pump operated at 30 μl/h.
Peptide Nomenclature
Peptides were designated by a serial number prefixed by a letter. The letters indicate the type of fragmentation as follows: K, lysyl endopeptidase; M, CNBr; D, endoproteinase Asp-N; X, Pfu N-acetyl deblocking aminopeptidase. The numbers in the designation do not correspond to the order of elution of the peptides in HPLC but rather to their positions in the protein sequence starting from the amino terminus.
Frontal Affinity Chromatography Technology
The purified lectin (2.4 mg) was dialyzed, dissolved in 0.1 m NaHCO3, pH 8.3, containing 0.5 m NaCl and 0.1 m lactose, and coupled to an N-hydroxysuccinimide-activated Sepharose 4 Fast Flow (2 ml) overnight at 4 °C. Uncoupled groups on the gel were then masked with 1 m ethanolamine-HCl, pH 8, overnight. The lectin-immobilized Sepharose gel was washed with TBS and packed into a miniature column (4 × 10 mm, 126 μl), and the column was connected to an HPLC system consisting of a pump (PU-2089, Jasco), fluorescence detector (FP-2020 Plus, Jasco), and data processing integrator (ChromNAV, Jasco).
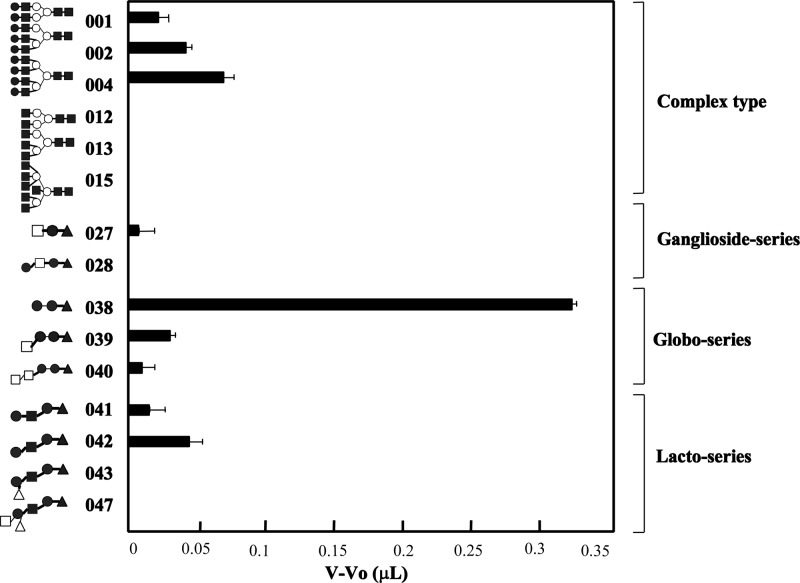
Fifteen PA-oligosaccharides (001, 002, 004, 012, 013, 015, 027, 028, 038–043, and 047) and PA-rhamnose (negative control) were applied manually to the affinity column (5 pmol each in 2 ml) at a flow rate of 250 μl/min and column temperature at 20 °C as described previously (19, 20). The front elution volume of the PA-oligosaccharides was detected by fluorescence detector measurements at 310 nm (excitation) and 380 nm (emission). The retardation volume (V − V0) of each PA-oligosaccharide (V) compared with the negative control (V0) was estimated by conversion from a bar graph. Analyses were performed in triplicate for each PA-oligosaccharide, and mean values were presented as bars.
KD Value of the Lectin Determined by Surface Plasmon Resonance
Gb3-HSA (50 μg/ml) dissolved in 10 mm sodium acetate, pH 4.5, was coupled to the carboxymethyl-dextran matrix of a sensor chip CM5 using a ligand coupling kit according to the manufacturer's instructions. The remaining uncoupled activated residues on the chip were masked with 1 m neutralized ethanolamine, pH 8. Various concentrations of purified lectin (0–100 μm) in HEPES-buffered saline were applied to the surface of a Gb3-HSA-conjugated sensor chip using an auto-sampler. The association and dissociation of the lectin to the immobilized Gb3-HSA on the chip were analyzed for 2 min at a flow rate of 20 μl/min at 25 °C (40). The sensor chip was subjected to association analysis, and the bound lectin was washed with HEPES-buffered saline and then removed from the sensor chip surface by 200 mm melibiose containing HEPES-buffered saline. The transition from association to dissociation of the lectin to the porcine stomach mucin on the sensor chip was monitored optically as a sensorgram. The dissociation constant estimated by the association rate (ka) and dissociation rate (kd) of each concentrated lectin was determined using the BIAevaluation software program, version 3.0 (GE Healthcare).
Cell Viability and Cytotoxicity Assays
Raji cells and K562 cells were maintained in RPMI 1640 medium supplemented with heat-inactivated fetal calf serum (10%, v/v), penicillin (100 IU/ml), and streptomycin (100 μg/ml) at 37 °C in an atmosphere of 95% air, 5% CO2. Cytotoxic activity and cell growth following treatment with various concentrations of the lectin (0–50 μg/ml) were determined by the cell counting kit-8 containing WST-8 (42–44, 46) and by trypan blue (0.5% (w/v) exclusion assay (45), respectively. To evaluate sugar inhibitory effects, sucrose, melibiose, and lactose (each 100 mm) were co-incubated with the lectin (20 μg/ml) for 24 h and then applied to the assay system. Cells (2 × 104, in 90 μl solution) were seeded into a 96-well flat-bottom plate and treated with various concentrations of the lectin (10 μl) for 24 h at 37 °C. The effect on cell growth was assayed by addition of WST-8 solution (10 μl) to each well and incubated for 4 h at 37 °C. The reduction in proportion of living cells was assayed by measurement of absorbance at 450 nm (reference, 600 nm) using the GLOMAX MultiDetection System (Promega).
Detection of Inversion or Perforation of Cell Membranes Following Lectin Treatment
Cells (2 × 105) were cultured in the presence of various concentrations (0–50 μg/ml) of the lectin for 24 h. Inversion and perforation of cell membranes were evaluated by treatment with fluorescein isothiocyanate (FITC)-conjugated anti-human annexin V rat monoclonal antibody and propidium iodide using the MEBCYTO apoptosis kit at 4 °C for 30 min. The reactions induced by addition of the lectin were detected by FACSCalibur (BD Biosciences) with a single laser emitting excitation at 488 nm (41–44, 47).
Statistical Analysis
Results of experiments are presented as the means ± S.E. Differences in means were evaluated by two-tailed Student's t test with p values <0.05 considered to be statistically significant.
RESULTS
Purification of Lectin from M. galloprovinicialis (“MytiLec”)
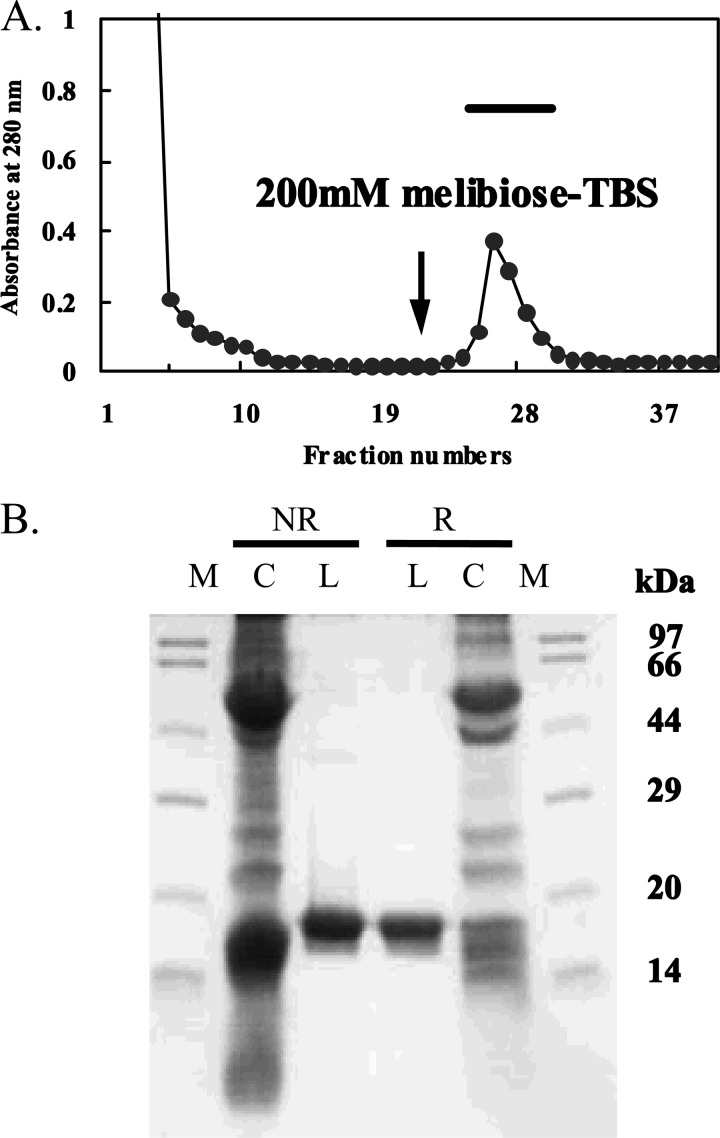
The crude supernatant (Sup 1) and the dialyzed extract (Sup 2) produced by addition of 100 mm galactose containing TBS to the precipitate both strongly agglutinated the trypsinized and glutaraldehyde-fixed rabbit erythrocytes. In affinity chromatography of Sup 1 applied to the melibiosyl-agarose column, a single peak appeared upon elution with TBS containing melibiose (Fig. 1A). SDS-PAGE showed the lectin as a single polypeptide with molecular mass of 17 kDa under reducing and nonreducing conditions (Fig. 1B). The purification of 100 g of fresh mussel tissue yielded 0.27 mg of lectin (Table 1). The 17-kDa lectin purified from M. galloprovincialis is hereafter designated as MytiLec. The hemagglutinating activity of MytiLec was not inhibited by the presence of 50 mm EDTA or 10 mm 2-mercaptoethanol (data not shown).
FIGURE 1.
Purification of MytiLec. A, extract from mussel (M. galloprovincialis) by TBS containing 100 mm d-Gal was extensively dialyzed and applied to a melibiose-conjugated agarose column (1 × 5 cm) equilibrated with TBS. MytiLec bound to the column was eluted with TBS containing 200 mm melibiose (arrow). B, SDS-PAGE pattern under reducing (R) and nonreducing (NR) conditions. Numbers at right indicate the molecular masses of marker proteins as follows: phosphorylase b (97 kDa), BSA (66 kDa), Ovalbumin (44 kDa), carbonic anhydrase (29 kDa), trypsin inhibitor (20 kDa), and lysozyme (14 kDa). M, molecular marker; C, crude extract; L, lectin.
TABLE 1.
Purification of MytiLec from Mytilus galloprovincialis
| Fraction | Titer | Volume | Total activitya | Protein concentration | Protein amount | Specific activityb | Purification ratioc | Recovery of activityd |
|---|---|---|---|---|---|---|---|---|
| ml | mg ml−1 | mg | -fold | % | ||||
| Crude extract obtained by TBS | 512 | 800 | 409,600 | 4.56 | 3,648 | 0.14 | 1 | 100 |
| Purified lectin | 8,192 | 10 | 81,920 | 0.92 | 9 | 910 | 6,500 | 20 |
| Crude extract obtained by galactose in TBS | 1,024 | 200 | 204,800 | 1.52 | 304 | 3.4 | 1 | 100 |
| Purified lectin | 8,192 | 11 | 99,112 | 0.4 | 4.4 | 1,862 | 547 | 48 |
a Total activity is calculated as titer × volume.
b Specific activity is calculated as titer/mg of protein.
c Purification ratio is calculated by comparing the value of specific activity on the crude extract versus purified lectin.
d Recovery of activity is calculated by comparing the value of total activity on the crude extract versus purified lectin.
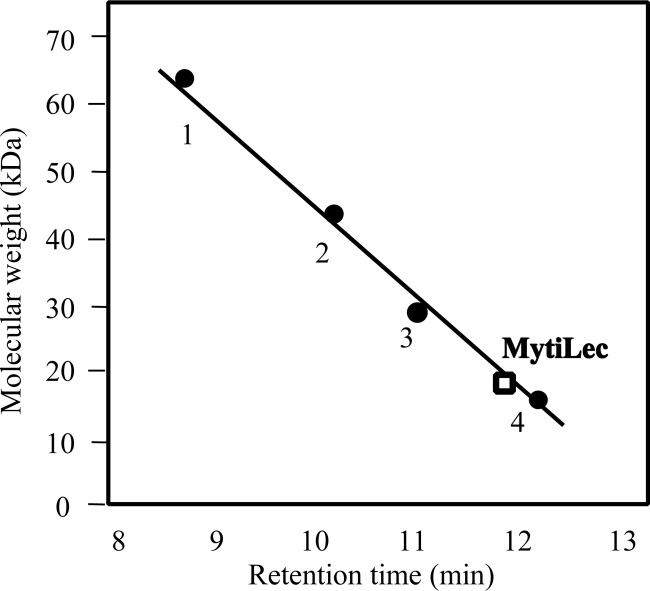
MytiLec Appears as a Monomer on GPC
MytiLec was eluted at a molecular mass of 17 kDa in TBS containing melibiose on a Superdex 75 column (Fig. 2), indicating that MytiLec existed as a monomer in the solution.
FIGURE 2.
Estimation of the molecular mass of MytiLec by gel permeation chromatography. A Superdex 75 column (1.0 × 30 cm) was equilibrated with TBS containing 50 mm melibiose. Molecular standards are shown in relation to elution position. Marker proteins: BSA (66 kDa) (1); ovalbumin (44 kDa) (2); carbonic anhydrase (29 kDa) (3), and lysozyme (14 kDa) (4).
Sugar Binding Specificity and Hemagglutination Inhibition Assay
The sugar binding specificity of MytiLec is summarized in Table 2. The hemagglutinating activity was markedly inhibited by the addition of monosaccharides such as d-Gal (3.13 mm) and d-GalNAc (1.56 mm) and of galactosides such as melibiose (1.56 mm), methyl α-d-galactopyranoside (3.13 mm), and methyl α-N-acetyl d-galactosaminide (1.56 mm). The activity was inhibited moderately by the addition of d-talose (25 mm), the C-2 epimer of d-Gal. In contrast, d-gulose, the C-3 epimer of d-Gal, did not inhibit the activity even at concentrations of >50 mm. β-d-Galactosides such as lactose, methyl β-d-galactopyranoside, and methyl β-N-acetyl d-galactosaminide, had less inhibitory effect than did α-d-galactosides (Table 2).
TABLE 2.
Saccharide and glycoprotein specificity of MytiLec
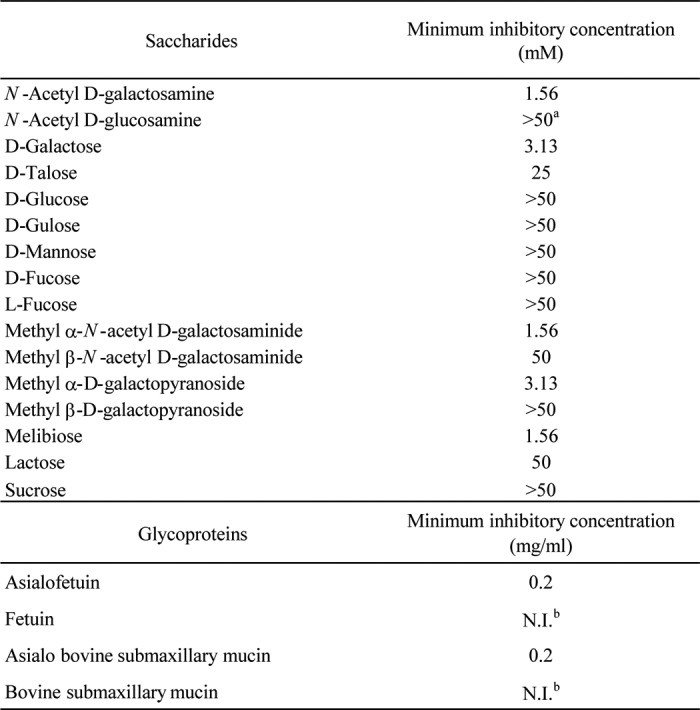
a Inhibition was not observed even at 100 mM.
b Fetuin and bovine submaxillary mucin had no inhibitory effect even at 2 mg/ml. The titer of MytiLec was previously diluted to 16.
Primary Structure of MytiLec
Edman degradation of 200 pmol of MytiLec gave very minor signals of XPNETKLVLXQDXHD, but the yield of phenylthiohydantoin-amino acid derivatives did not match the amount loaded on the sequencer. The lectin was therefore presumed to have a blocked amino terminus. The minor sequence observed was found later as an internal sequence. The acid-labile Asp-Pro linkage may have been partially cleaved during the RP-HPLC process under acidic conditions. The amino acid composition of MytiLec is shown in Table 3. No significant cysteine residue signal was detected. Because ∼4 Met, 14 Lys, and 21 Asx residues per molecule were found in the amino acid composition, MytiLec purified by RP-HPLC was cleaved by enzymatic (lysyl endopeptidase and endoproteinase Asp-N) and chemical (CNBr) agents to generate peptide fragments for structural analysis. The proven 149-residue sequence of MytiLec is summarized in Fig. 4. This sequence was established primarily through the overlapping of peptide sequences obtained by Edman degradation of peptide fragments generated from MytiLec using the above agents.
TABLE 3.
Amino acid components of MytiLec
| Amino acids | Residue/mola |
|---|---|
| Asx | 20.9 (11/9)b |
| Thr | 6.9 (7) |
| Ser | 6.8 (7) |
| Glx | 7.9 (5/2) |
| Gly | 12.7 (12) |
| Ala | 9.4 (9) |
| Val | 10.3 (11) |
| Met | 3.8 (4) |
| Ile | 5.9 (7) |
| Leu | 10.3 (10) |
| Tyr | 4.1 (4) |
| Phe | 9.4 (9) |
| Lys | 14.3 (15) |
| His | 11.8 (12) |
| Arg | 5.5 (5) |
| Cys | NDc (0) |
| Pro | 8.8 (9) |
| Trp | 0.5 (1) |
a Values are expressed as residues/molecule.
b Values in parentheses are taken from the sequence.
c ND means not detected.
FIGURE 4.
Proven sequence of MytiLec. Sequences determined by Edman degradation of specific peptides (indicated by italic type) are shown by one-letter code below the summary sequence (top line). Molecular mass values obtained from MALDI-TOF and FT-ICR MS are indicated in parentheses. Uppercase letters, peptide sequences proven by Edman degradation. Lowercase letters, sequences tentatively identified or deduced from MS. m′, homoserine lactone. Dashes, sequences not identified by the protein sequencer.
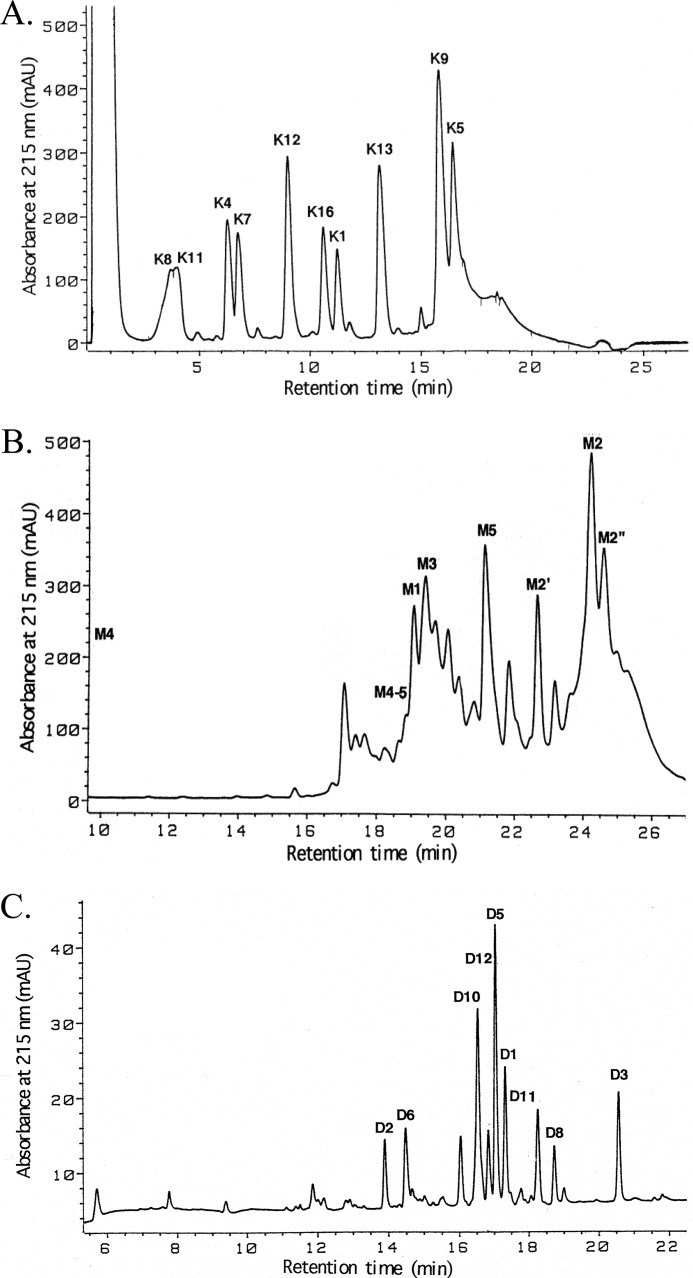
Lysyl Endopeptidase Digestion
Nine major peptides were recovered from a digest of MytiLec (2 nmol) upon separation by RP-HPLC on an Aquapore RP-300 column (Fig. 3A). Peptide K1 was presumed to be derived from the blocked amino terminus because it was refractory to Edman degradations. Sequence analyses of other isolated K peptides yielded a 115-residue sequence and provided overlaps from M2 to M5. A linear sequence of 132 residues (residues 18–149) was thereby established (Fig. 4).
FIGURE 3.
Elution (HPLC) profiles of peptides generated by enzymatic digestion and chemical cleavage of MytiLec. A, Achromobacter protease I digest on an Aquapore RP-300 column. B, CNBr cleavage on a Superspher Select B column. C, Asp-N digest on a Mightysil RP-18 column. Peptides were eluted by a gradient of acetonitrile into dilute aqueous TFA.
CNBr Cleavage
CNBr-cleaved fragments of MytiLec (2 nmol) were separated by RP-HPLC on a Superspher Select B column (Fig. 3B). The smallest fragment, M4, was involved in the pass-through peak. Five major peptides, M1 to M5, were isolated along with some overlap fragments and two fragments generated by an acid-labile Asp-Pro bond cleavage. Isolated fragments were subjected to Edman degradation (Fig. 4) and mass spectrometry (Table 4). Peak M1 (observed molecular mass 4292.2) yielded no phenylthiohydantoin signal, indicating the presence of a blocked amino terminus. Sequence analyses of other isolated fragments yielded 100 amino acid residues showing the elution positions M2′, M2″, and M4–5 in Fig. 4.
TABLE 4.
MALDI-TOF MS analysis of derived peptides of MytiLec
| Peptides | Residue no. | Observeda | Calculatedb |
|---|---|---|---|
| M1 | 1–39 | 4292.2 | 4294.8* |
| M2 | 40–91 | 5925.3 | 5928.7* |
| M2′ | 40–70 | 3504.2 | 3506 |
| M2″ | 71–91 | 2441.8 | 2441.8 |
| M3 | 92–123 | 3495.9 | 3493.8* |
| M4 | 124–131 | 804.4 | 804.4 |
| M4–5 | 124–149 | 2987.5 | 2984.6 |
| M5 | 132–149 | 2182.3 | 2181.2 |
| K1 | 1–6 | 764.6 | 764.5 |
| K4 | 13–17 | 641.6 | 641.4 |
| K5 | 18–54 | 4309.2 | 4310 |
| K7 | 61–68 | 820.7 | 820.5 |
| K8 | 69–76 | 871.8 | 871.4 |
| K9 | 77–101 | 3067.1 | 3065.6 |
| K11 | 105–109 | 643.6 | 643.4 |
| K12 | 110–127 | 1855 | 1854.9 |
| K13 | 128–139 | 1363 | 1362.7 |
| K16 | 144–149 | 748.6 | 748.5 |
| D1 | 1–25 | 2680.4 | 2679.4 |
| D2 | 26–33 | 898.7 | 898.5 |
| D3 | 34–43 | 1385.8 | 1385.6 |
| D5 | 47–69 | 2491.2 | 2490.3 |
| D6 | 70–81 | 1392.1 | 1390.7 |
| D8 | 85–91 | 823.5 | 823.4 |
| D10 | 96–125 | 3213.3 | 3212.6 |
| D11 | 126–141 | 1849 | 1848 |
| D12 | 142–149 | 992 | 991.6 |
| Pfu-X1 | 6–25 | 2063.3 | 2061.5 |
a Values were calculated for MH+ from multiple charged signals observed.
b Data were calculated as monoisotopic mass or average mass (*).
Endoproteinase Asp-N Digestion and Subdigestion by Pfu Deblocking Aminopeptidase
MytiLec (2 nmol) was digested with endoproteinase Asp-N, and nine major peptides were separated by RP-HPLC on a Mightysil RP-18 column (Fig. 3C). Mass spectral analysis of each peak revealed that the recovered peptides covered the whole protein except for two tripeptides and one tetrapeptide expected from the completed portion of the molecule (Table 4). The amino-terminally blocked peptide D1 (observed molecular mass 2680.4) was subdigested by Pfu-deblocking aminopeptidase. Five peptides, including undigested ones, were separated by RP-HPLC (data not shown), and the peptide that had the largest molecular mass among the deblocked peptides was subjected to sequence analysis. Results provided the 12-residue extension toward the amino terminus of the protein that included K4 (X1 in Fig. 4).
Amino-terminal Sequence and the Blocking Group
The post-source decay spectrum of K1 (M+ = 764.5 by ESI/FT-ICR MS) by MALDI-TOF MS indicated that the carboxyl-terminal sequence of the peptide was likely to be -Phe-Leu (or Ile)-Leu (or Ile)-Lys. The spectrum indicated the presence of two Thr residues, with and without an acetyl group, suggesting that the amino terminus of MytiLec was likely to be acetyl-Thr. The amino acid composition of K1 included two Thr residues (∼400 pmol) and one residue each of Phe, Leu, Ile, and Lys (∼200 pmol each). To confirm the sequence, K1 was treated with concentrated (12 n) HCl to remove the acetyl group from the amino-terminal Thr by N-O shift reaction and subjected to Edman degradation. The analysis revealed the sequence Thr-Thr-Phe-Leu-Ile-Lys (Fig. 4). The amino-terminal Lys of the X1 peptide generated by the Pfu-deblocking aminopeptidase appeared to be the carboxyl-terminal Lys of K1 because the calculated mass of acetyl-Thr1 through Asn25 was consistent with the observed mass of the amino-terminal peptide D1 (Table 4).
Complete Primary Structure of MytiLec, with Novel Triple Tandem Repeat
The complete primary structure of MytiLec, consisting of 149 amino acids, was completed by the sequence of overlapping peptides (Fig. 4). The precise molecular mass of the whole protein was determined to be 16,812.59 Da by ESI/FT-ICR MS (data not shown). This value was in good agreement with the calculated value of 16,823.22 Da for the lectin. The amino-terminal amino group of Thr was blocked by an acetyl group. This very novel primary structure was elucidated by searches of the database in comparison with the structures of previously reported animal lectins. MytiLec had a triple tandem-repeat structure with three domains each consisting of ∼50 amino acids with 50% homology (Fig. 5). Basic amino acids such as Lys and His (blue) were located on the whole polypeptide, whereas acidic amino acids such as Asp and Glu (red) were located on the carboxyl-terminal side within the domain.
FIGURE 5.

Amino acid sequence homology of the internal tandem-repeat domains of MytiLec. A, boxes indicate the common (homologous) amino acid residues within the repeat domains. Common residues at bottom summarizes the common residues in the three domains. Uppercase letters, residues identical in all three domains. Lowercase letters, residues identical in two of the domains. B, residue numbers of the polypeptide are shown. Acidic and basic amino acids are indicated as Acidic a.a. and Basic a.a., respectively.
Glycan Binding Profile of MytiLec by FACT
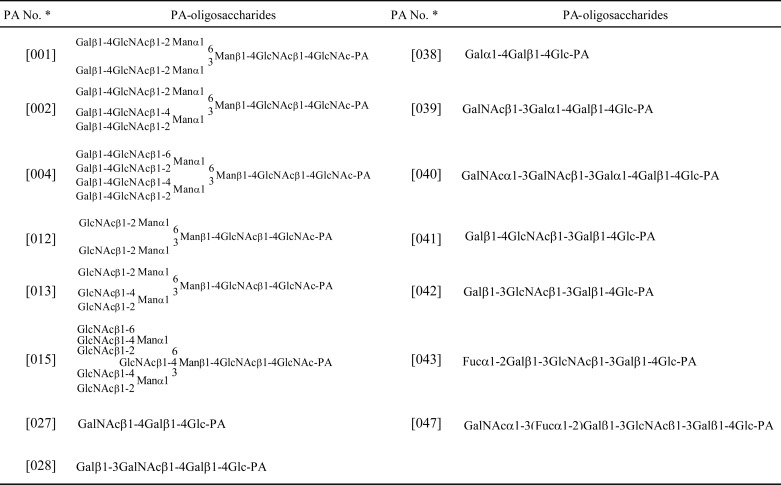
The determination of the glycan-binding profile of MytiLec used 15 PA-oligosaccharides as shown in Table 5 and Fig. 6. MytiLec had a strong affinity with globotriose (Gb3; 038). MytiLec showed a weak affinity with branched type 2 N-acetyllactosamine chains (Galβ1–4GlcNAc) attached to the core structure, e.g. bi-antennary; 001 = tri-antennary; 002 < tetra-antennary; 004. MytiLec bound weakly to α-N-acetylgalactosaminyl (040) and moderately to type 1 (Galβ1–3GlcNAc) (041) and type 2 (Galβ1–4GlcNAc) (042) lactosamine. The glycan-binding profile was consistent with the finding that MytiLec was selectively purified by a melibiosyl-agarose column involving α-galactoside (Fig. 6).
TABLE 5.
List of PA-oligosaccharides for FACT analysis
a Numbering of PA oligosaccharides is followed by the product codes of Takara Bio Inc.
FIGURE 6.
Glycan-binding profile of MytiLec obtained by FACT analysis. The numbers on the vertical axis correspond to the oligosaccharide numbers in the list of PA-glycans shown in Table 5 and used in the text. Horizontal axis, difference in relative intensity between the elution front volume of each PA-oligosaccharide (V) and PA-rhamnose (Vo). Error bars, S.E. (n = 3).
Association and Dissociation Rates of MytiLec
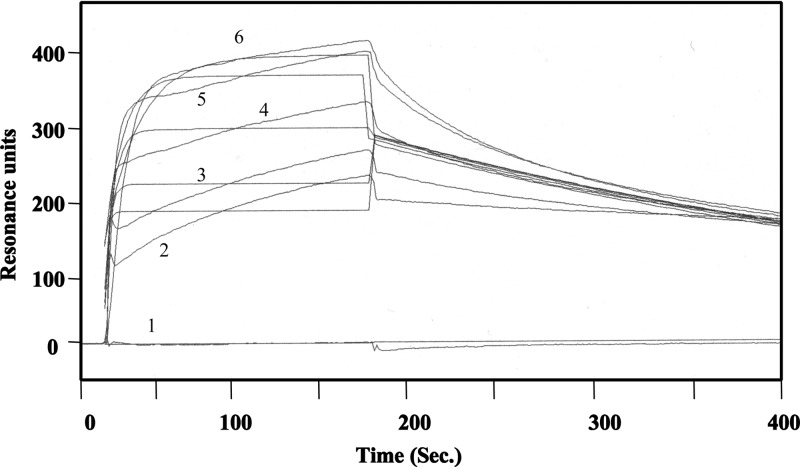
Gb3-HSA was immobilized on the CM5 sensor chip at 4300 resonance units, and several concentrations of MytiLec were applied as analytes to determine the dissociation constant of the lectin using a Biacore 3000. SPR analysis indicated that free MytiLec used as an analyte bound to the immobilized Gb3-HSA in a dose-dependent manner (Fig. 7). The ka, kd, and KD values of MytiLec were calculated as 1.4 × 106 m−1 s−1, 3.1 × 10−2 s−1, and 2.2 × 10−8 m, respectively. These results indicate that MytiLec associates quickly and dissociates slowly from the neoglycoprotein with strong binding affinity.
FIGURE 7.
Kinetic analysis by SPR. MytiLec was applied to a CM5 sensor chip coupled with Galα1–4Galβ1–4Glc-HSA at 6.8 ng/mm2. MytiLec was applied to a asialofetuin-conjugated sensor chip at 20 μl/min for 2.5 min. MytiLec concentrations (red lines from top to bottom): 800 (6), 400 (5), 200 (4), 100 (3), 50 (2), and 0 (1) nm. The chip was washed with TBS for 2.5 min. Vertical axis, resonance units (indicating association and dissociation of the analyte). Kinetics were analyzed using the BIAevaluation software program, version 3.0 (GE Healthcare).
MytiLec Signal Transduction Reduces Cell Viability
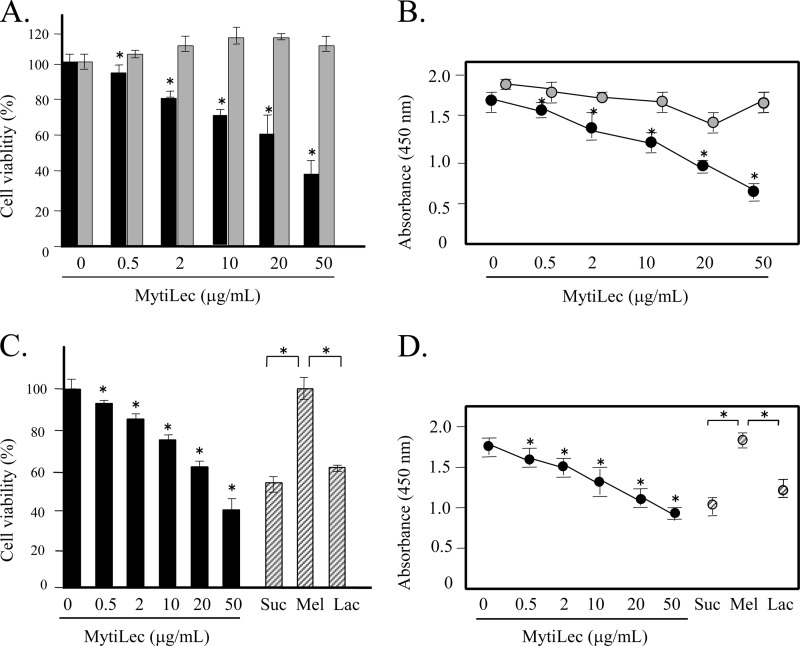
In view of the finding that MytiLec bound strongly to Gb3, the lectin was added experimentally to the culture medium of Raji cells, which abundantly express Gb3 on the cell membrane. Various concentrations of MytiLec were incubated with 105 cells/ml for 24 h, and cell viability and the ratio of living cells were measured by trypan blue assay and WST-8 assay, respectively. As the concentration of MytiLec increased from 0.5 to 50 μg/ml, cell viability declined (Fig. 8A, black bars). K562 cells (negative control) were unaffected by MytiLec addition (Fig. 8A, gray bars). The living cell ratio (Fig. 8B) was correlated with the viability results. These findings indicate that MytiLec effectively inhibited the viability of Burkitt lymphoma cells.
FIGURE 8.
Glycan-dependent reduction of viability of Burkitt lymphoma Raji cells by MytiLec. Cell viability and numbers of living cells were determined using trypan blue exclusion assay (A and C) and WST-8 (B and D), respectively. A and B, Raji cells were treated with various concentrations (0–50 μg/ml) of MytiLec. Black bars (or circles) and gray bars (or circles) indicate the viability (or absorbance) of Raji cells and erythroleukemia K562 cells (negative control), respectively. C and D, blocking by various added saccharides (100 mm each) of the reduction of cell viability caused by MytiLec. Suc, sucrose; Mel, melibiose; Lac, lactose. Black bars (or circles) and shaded bars (or circles) indicate the viability (or absorbance) of Raji cells and Raji cells added with saccharides, respectively. Error bars, S.E. (n = 3).
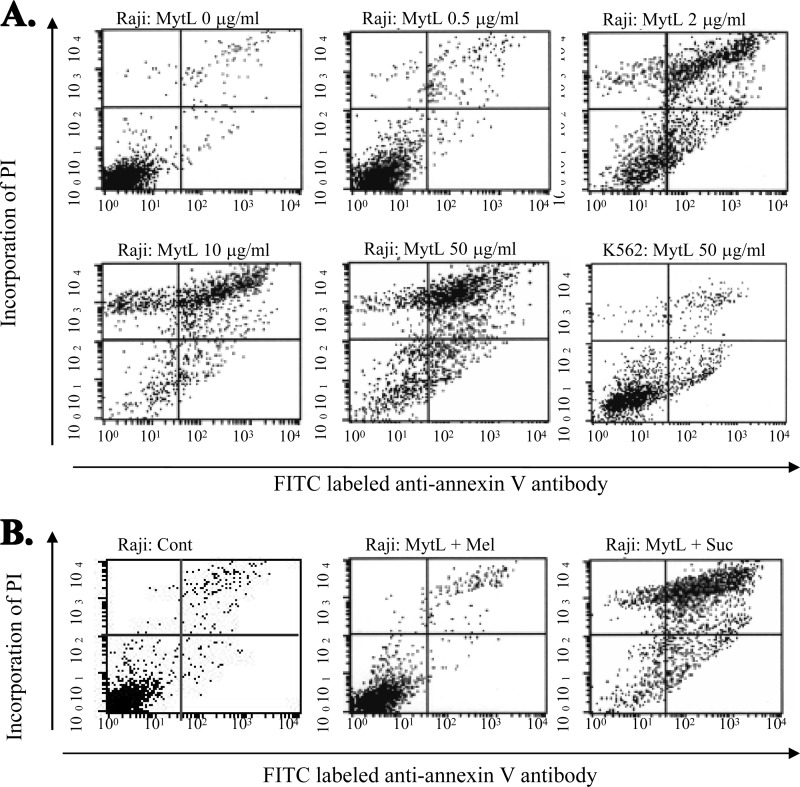
The possible blocking of the inhibitory effect of MytiLec on the viability by the co-presence of sugars was investigated. The β-galactosides sucrose and lactose had no such blocking effect. In contrast, the co-presence of melibiose (a α-galactoside, like Gb3) completely blocked the cytotoxic effect of MytiLec (Fig. 8, C and D). This finding indicates that a specific glycan structure such as Gb3 on Raji cells was essential for the cytotoxic effect of MytiLec. FACS analysis revealed that MytiLec treatment was associated with deleterious biological phenomena such as cell membrane inversion and loss of membrane integrity (Fig. 9). The horizontal axes in Fig. 9 show the binding of FITC-labeled anti-annexin V antibody, and the vertical axe shows the incorporation of propidium iodide. As the MytiLec concentration increased, the population of anti-annexin V-positive Raji cells in the histogram shifted to the right, indicating the occurrence of cell membrane inversion. The incorporation of propidium iodide also indicated penetration of the cell membrane following MytiLec treatment because the cells shifted to the upper part of the histogram (Fig. 9A, Raji: MytL 0–50 μg/ml). Such effects were not observed for K562 cells treated with MytiLec (Fig. 9A, K562: MytL 50 μg/ml). The sensitivity of both annexin V and propidium iodide to MytiLec was completely blocked by addition of the α-galactoside melibiose (Fig. 9B, Raji + Mel).
FIGURE 9.
Detection of annexin V and incorporation of propidium iodide (PI) in MytiLec-treated Raji cells as analyzed by FACSCalibur. Horizontal axes, binding of FITC-labeled anti-annexin V antibody. Phosphatidylserine externalization and propidium iodide incorporation were detected using the MEBCYTO apoptosis kit by FACScan. A, Raji and K562 cells were treated with various concentrations (0–50 μg/ml) of MytiLec (MytL) as indicated for 30 min at 4 °C. B, blocking of the cytotoxic activity of MytiLec (10 μg/ml) by saccharide (100 mm) addition to Raji cells. Cont, PBS; Mel, melibiose; Suc, sucrose.
DISCUSSION
The complete primary structure determined for MytiLec, a lectin isolated from the mussel M. galloprovincialis, is unique among the structures of other known animal lectins. It consists of 149 amino acids without similarity to other known structures. MytiLec has a triple tandem-repeat motif of 50 amino acids. The three motifs show >50% similarity with each other. The basic amino acid residues of Lys, His, and Arg are highly conserved throughout the domains of the motifs, whereas the acidic amino acid residues of Asp and Glu are located mainly in the carboxyl-terminal side. Other characteristic features of the MytiLec structure are one Trp, 12 His, and no Cys residues involved in the polypeptide. The high number of conserved amino acid residues (His, Asp, and Arg) in the polypeptide is interesting in that these residues are found as essential carbohydrate-binding amino acids in many lectins (48–52). Acetylation at the amino-terminal Thr was the only observed post-translational modification in MytiLec; there was neither glycosylation nor phosphorylation. Results from gel permeation chromatography indicate that MytiLec is present as a monomer, suggesting that each of its polypeptide motifs has hemagglutinating activity. It is interesting that a motif consisting of 50 amino acids could have carbohydrate binding ability; well known animal lectin families such as galectins and C-type lectins require >130 amino acids to function as a carbohydrate-recognition domain (5, 51). Structural biological studies of MytiLec will provide additional information regarding the glycan binding properties of the polypeptide subdomains. The highly novel primary structure of MytiLec has no known homologues at present, but we anticipate that our finding will lead to future studies that reveal such homologues. In analogy, the primary structure of d-galactoside-binding lectin isolated from SUEL had no structural homologues when we first reported it in 1991 (11); however, >1000 structural homologues with SUEL-type lectin domains have been found during the 20 years since then. This structure has been observed even in the lectin domain of a neurotoxin receptor (latrophilin-1) in mammalian brain (52, 53) and in plant β-galactosidase (54). As an example of a new structural domain in animal lectins, MytiLec is of interest in and will promote the field of glycobiology.
Our next interest will be a survey of α-galactosides in other animal species. An α-galactoside was specifically recognized by MytiLec in FACT analysis, although the occurrence of these sugars in mussels is not yet clear. Certain Gal-containing oligosaccharides in glycoproteins have been recently found in some mollusks by mass spectrometric analysis (55–57). MytiLec was obtained together with the haptenic saccharide Gal and therefore bound to mussel tissues. Isolation of the endogenous ligands of MytiLec will help elucidate the physiological roles of Gal-binding lectins isolated from other mollusk species.
FACT analysis showed that MytiLec specifically recognizes Gb3. Other lectins in marine organisms have been previously found to recognize α-galactosides (11, 12, 23). Because of the characteristic glycan binding property of MytiLec in recognizing the α-galactoside Gb3, it selectively aggregated and directly killed human Burkitt lymphoma Raji cells, which express Gb3 ceramide in the glycosphingolipid-enriched microdomain of the cell membrane through glycan-lectin interaction. FACS analysis indicated that MytiLec was associated with late-stage apoptosis and induced both cell membrane inversion and the loss of membrane integrity. It will be interesting to investigate how transduction of the lectin signal kills cells that specifically express Gb3 in glycosphingolipid-enriched microdomain. We showed recently that another Gb3-binding lectin with a SUEL-type lectin domain, isolated from catfish eggs, reduced the expression of mRNA coding a multidrug-resistant transporter in Raji cells (58). MytiLec and SAL both have a triple-tandem structure but differ in terms of multivalency; MytiLec is structured as a monomer (containing three carbohydrate-recognition domains in total), whereas SAL is a trimer (containing nine carbohydrate-recognition domains in total) under physiological conditions. If it is found that the same ligand is recognized by lectins that have a different affinity constant and multimerization, we could hypothesize that independent signal pathways are being activated, with results differing from those of other regulatory pathways in the same cells. Subsequent studies will determine the targeting signal transduction molecules that are stimulated by MytiLec and tissue localization of MytiLec during mussel development. Various types of molecules are known to modulate signal transduction from glycosphingolipid-enriched microdomain in cell membranes. Molecular recognition between MytiLec and Gb3 may play an important role in regulating the fate of cells.
Acknowledgments
We thank the City of Yokohama “Blue Carbon Project” and Yachiyo Engineering Co., Ltd., for their support. Naoko Masuda and Chihiro Iwahara are specially thanked for assistance with experiments. We thank Dr. Stephen Anderson for the English editing of the manuscript.
This work was supported in part by grants-in-aid for scientific research from the Japan Society for the Promotion of Science and Japanese Association for Marine Biology from the Ministry of Education, Culture, Sports, Science, and Technology Japan.
The protein sequence data reported in this paper will appear in the UniProt Knowledgebase under the accession number B3EWR1 for MytiLec.
- SUEL
- sea urchin egg lectin
- FACT
- frontal affinity chromatography technology
- FT-ICR MS
- Fourier-transform ion cyclotron resonance mass spectrometry
- Gb3
- globotriaosylceramide
- GPC
- gel permeation chromatography
- MytiLec
- Mytilus galloprovincialis α-d-galactose-binding lectin
- PA
- pyridylamino
- Sup
- supernatant
- HSA
- human serum albumin
- SPR
- surface plasmon resonance.
REFERENCES
- 1. Kinoshita S., Wang N., Inoue H., Maeyama K., Okamoto K., Nagai K., Kondo H., Hirono I., Asakawa S., Watabe S. (2011) Deep sequencing of ESTs from nacreous and prismatic layer producing tissues and a screen for novel shell formation-related genes in the pearl oyster. PLoS ONE 6, e21238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Venier P., De Pittà C., Bernante F., Varotto L., De Nardi B., Bovo G., Roch P., Novoa B., Figueras A., Pallavicini A., Lanfranchi G. (2009) MytiBase. A knowledge base of mussel (M. galloprovincialis) transcribed sequences. BMC Genomics 10, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Venier P., Varotto L., Rosani U., Millino C., Celegato B., Bernante F., Lanfranchi G., Novoa B., Roch P., Figueras A., Pallavicini A. (2011) Insights into the innate immunity of the Mediterranean mussel Mytilus galloprovincialis. BMC Genomics 12, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kim Y. M., Park K. I., Choi K. S., Alvarez R. A., Cummings R. D., Cho M. (2006) Lectin from the Manila clam Ruditapes philippinarum is induced upon infection with the protozoan parasite Perkinsus olseni. J. Biol. Chem. 281, 26854–26864 [DOI] [PubMed] [Google Scholar]
- 5. Pales Espinosa E., Perrigault M., Allam B. (2010) Identification and molecular characterization of a mucosal lectin (MeML) from the blue Mytilus edulis and its potential role in particle capture. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 156, 495–501 [DOI] [PubMed] [Google Scholar]
- 6. Tasumi S., Vasta G. R. (2007) A galectin of unique domain organization from hemocytes of the Eastern oyster (Crassostrea virginica) is a receptor for the protistan parasite Perkinsus marinus. J. Immunol. 179, 3086–3098 [DOI] [PubMed] [Google Scholar]
- 7. Gorbushin A. M., Iakovleva N. V. (2011) A new gene family of single fibrinogen domain lectins in Mytilus. Fish Shellfish Immunol. 30, 434–438 [DOI] [PubMed] [Google Scholar]
- 8. Li C., Yu S., Zhao J., Su X., Li T. (2011) Cloning and characterization of sialic acid binding lectins (SABL) from Manila clam Venerupis philippinarum. Fish Shellfish Immunol. 30, 1202–1206 [DOI] [PubMed] [Google Scholar]
- 9. Chen J., Xiao S., Yu Z. (2011) F-type lectin involved in defense against bacterial infection in the pearl oyster (Pinctada martensii). Fish Shellfish Immunol. 30, 750–754 [DOI] [PubMed] [Google Scholar]
- 10. Belogortseva N. I., Molchanova V. I., Kurika A. V., Skobun A. S., Glazkova V. E. (1998) Isolation of characterization of new GalNAc/Gal-specific lectin from the sea mussel Crenomytilus grayanus. Comp. Biochem. Physiol. C Pharmacol. Toxicol. Endocrinol. 119, 45–50 [DOI] [PubMed] [Google Scholar]
- 11. Ozeki Y., Matsui T., Suzuki M., Titani K. (1991) Amino acid sequence and molecular characterization of a d-galactoside-specific lectin purified from sea urchin (Anthocidaris crassispina) eggs. Biochemistry 30, 2391–2394 [DOI] [PubMed] [Google Scholar]
- 12. Naganuma T., Ogawa T., Hirabayashi J., Kasai K., Kamiya H., Muramoto K. (2006) Isolation, characterization, and molecular evolution of a novel pearl shell lectin from a marine bivalve, Pteria penguin. Mol. Divers. 10, 607–618 [DOI] [PubMed] [Google Scholar]
- 13. Blixt O., Head S., Mondala T., Scanlan C., Huflejt M. E., Alvarez R., Bryan M. C., Fazio F., Calarese D., Stevens J., Razi N., Stevens D. J., Skehel J. J., van Die I., Burton D. R., Wilson I. A., Cummings R., Bovin N., Wong C.-H., Paulson J. C. (2004) Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 17033–17038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Angeloni S., Ridet J. L., Kusy N., Gao H., Crevoisier F., Guinchard S., Kochhar S., Sigrist H., Sprenger N. (2005) Glycoprofiling with micro-arrays of glycoconjugates and lectins. Glycobiology 15, 31–41 [DOI] [PubMed] [Google Scholar]
- 15. Tateno H., Uchiyama N., Kuno A., Togayachi A., Sato T., Narimatsu H., Hirabayashi J. (2007) A novel strategy for mammalian cell surface glycome profiling using lectin microarray. Glycobiology 17, 1138–1146 [DOI] [PubMed] [Google Scholar]
- 16. Hirabayashi J. (2008) Concept, strategy, and realization of lectin-based glycan profiling. J. Biochem. 144, 139–147 [DOI] [PubMed] [Google Scholar]
- 17. Kasai K., Ishii S. (1978) Affinity chromatography of trypsin and related enzymes. V. Basic studies of quantitative affinity chromatography. J. Biochem. 84, 1051–1060 [DOI] [PubMed] [Google Scholar]
- 18. Kasai K., Ishii S. (1978) Studies on the interaction of immobilized trypsin and specific ligands by quantitative affinity chromatography. J. Biochem. 84, 1061–1069 [DOI] [PubMed] [Google Scholar]
- 19. Hirabayashi J., Hashidate T., Arata Y., Nishi N., Nakamura T., Hirashima M., Urashima T., Oka T., Futai M., Muller W. E., Yagi F., Kasai K. (2002) Oligosaccharide specificity of galectins. A search by frontal affinity chromatography. Biochim. Biophys. Acta 1572, 232–254 [DOI] [PubMed] [Google Scholar]
- 20. Hirabayashi J., Arata Y., Kasai K. (2003) Frontal affinity chromatography as a tool for elucidation of sugar recognition properties of lectins. Methods Enzymol. 362, 353–368 [DOI] [PubMed] [Google Scholar]
- 21. Kawsar S. M., Fujii Y., Matsumoto R., Ichikawa T., Tateno H., Hirabayashi J., Yasumitsu H., Dogasaki C., Hosono M., Nitta K., Hamako J., Matsui T., Ozeki Y. (2008) Isolation, purification, characterization and glycan-binding profile of a d-galactoside specific lectin from the marine sponge, Halichondria okadai. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 150, 349–357 [DOI] [PubMed] [Google Scholar]
- 22. Kawsar S. M., Takeuchi T., Kasai K., Fujii Y., Matsumoto R., Yasumitsu H., Ozeki Y. (2009) Glycan-binding profile of a d-galactose binding lectin purified from the annelid, Perinereis nuntia ver. vallata. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 152, 382–389 [DOI] [PubMed] [Google Scholar]
- 23. Kawsar S. M., Matsumoto R., Fujii Y., Matsuoka H., Masuda N., Chihiro I., Yasumitsu H., Kanaly R. A., Sugawara S., Hosono M., Nitta K., Ishizaki N., Dogasaki C., Hamako J., Matsui T., Ozeki Y. (2011) Cytotoxicity and glycan-binding profile of a d-galactose-binding lectin from the eggs of a Japanese sea hare (Aplysia kurodai). Protein J. 30, 509–519 [DOI] [PubMed] [Google Scholar]
- 24. Matsumoto R., Shibata T. F., Kohtsuka H., Sekifuji M., Sugii N., Nakajima H., Kojima N., Fujii Y., Kawsar S. M., Yasumitsu H., Hamako J., Matsui T., Ozeki Y. (2011) Glycomics of a novel type-2 N-acetyllactosamine-specific lectin purified from the feather star, Oxycomanthus japonicus (Pelmatozoa: Crinoidea). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 158, 266–273 [DOI] [PubMed] [Google Scholar]
- 25. Matsumoto R., Fujii Y., Kawsar S. M., Kanaly R. A., Yasumitsu H., Koide Y., Hasan I., Iwahara C., Ogawa Y., Im C. H., Sugawara S., Hosono M., Nitta K., Hamako J., Matsui T., Ozeki Y. (2012) Cytotoxicity and glycan-binding properties of an 18-kDa lectin isolated from the marine sponge Halichonderia okadai. Toxins 4, 323–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gourdine J. P., Cioci G., Miguet L., Unverzagt C., Silva D. V., Varrot A., Gautier C., Smith-Ravin E. J., Imberty A. (2008) High affinity interaction between a bivalve C-type lectin and a biantennary complex-type N-glycan revealed by crystallography and microcalorimetry. J. Biol. Chem. 283, 30112–30120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. (1985) Measurement of protein using bicinchoninic acid. Anal. Biochem. 150, 76–85 [DOI] [PubMed] [Google Scholar]
- 28. Wiechelman K. J., Braun R. D., Fitzpatrick J. D. (1988) Investigation of the bicinchoninic acid protein assay. Identification of the groups responsible for color formation. Anal. Biochem. 175, 231–237 [DOI] [PubMed] [Google Scholar]
- 29. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 30. Masaki T., Tanabe M., Nakamura K., Soejima M. (1981) Studies on a new proteolytic enzyme from Achromobacter lyticus M497-1. I. Purification and some enzymatic properties. Biochim. Biophys. Acta 660, 44–50 [DOI] [PubMed] [Google Scholar]
- 31. Gross E. (1967) Cleavage of peptide chains. The cyanogen bromide reaction. Methods Enzymol. 11, 238–255 [Google Scholar]
- 32. Tahirov T. H., Oki H., Tsukihara T., Ogasahara K., Yutani K., Ogata K., Izu Y., Tsunasawa S., Kato I. (1998) Crystal structure of methionine aminopeptidase from hyperthermophile, Pyrococcus furiosus. J. Mol. Biol. 284, 101–124 [DOI] [PubMed] [Google Scholar]
- 33. Titani K., Narita K. (1964) Amino acid sequences of 18 peptides isolated from the tryptic hydrolysate of Baker's yeast cytochrome c. J. Biochem. 56, 241–256 [DOI] [PubMed] [Google Scholar]
- 34. Simpson R. J., Neuberger M. R., Liu T. Y. (1976) Complete amino acid analysis of proteins from a single hydrolysate. J. Biol. Chem. 251, 1936–1940 [PubMed] [Google Scholar]
- 35. Hewick R. M., Hunkapiller M. W., Hood L. E., Dreyer W. J. (1981) A gas-liquid solid phase peptide and protein sequenator. J. Biol. Chem. 256, 7990–7997 [PubMed] [Google Scholar]
- 36. Altschul S. F., Lipman D. J. (1990) Protein database searches for multiple alignments. Proc. Natl. Acad. Sci. U.S.A. 87, 5509–5513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J., (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xu N., Huang Z.-H., Watson J. T., Gage D. A. (1997) Mercaptobenzothiazoles. A new class of matrices for laser desorption ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 8, 116–124 [Google Scholar]
- 39. Vukelić Z., Zamfir A. D., Bindila L., Froesch M., Peter-Katalinić J., Usuki S., Yu R. K. (2005) Screening and sequencing of complex sialylated and sulfated glycosphingolipid mixtures by negative ion electrospray Fourier transform ion cyclotron resonance mass spectrometry. J. Am. Soc. Mass Spectrom. 16, 571–580 [DOI] [PubMed] [Google Scholar]
- 40. Shinohara Y., Kim F., Shimizu M., Goto M., Tosu M., Hasegawa Y. (1994) Kinetic measurement of the interaction between an oligosaccharide and lectins by a biosensor based on surface plasmon resonance. Eur. J. Biochem. 223, 189–194 [DOI] [PubMed] [Google Scholar]
- 41. Kawano T., Sugawara S., Hosono M., Tatsuta T., Ogawa Y., Fujimura T., Taka H., Murayama K., Nitta K., (2009) Globotriaosylceramide-expressing Burkitt lymphoma cells are committed to early apoptotic status by rhamnose-binding lectin from catfish eggs. Biol. Pharm. Bull. 32, 345–353 [DOI] [PubMed] [Google Scholar]
- 42. Kawano T., Sugawara S., Hosono M., Tatsuta T., Nitta K. (2008) Alteration of gene expression induced by Silurus asotus lectin in Burkitt lymphoma cells. Biol. Pharm. Bull. 31, 998–1002 [DOI] [PubMed] [Google Scholar]
- 43. Sugawara S., Hosono M., Ogawa Y., Takayanagi M., Nitta K. (2005) Catfish egg lectin causes rapid activation of multidrug resistance 1 P-glycoprotein as a lipid translocase. Biol. Pharm. Bull. 28, 434–441 [DOI] [PubMed] [Google Scholar]
- 44. Sugawara S., Sasaki S., Ogawa Y., Hosono M., Nitta K., (2005) Catfish (Silurus asotus) lectin enhances the cytotoxic effects of doxorubicin. Yakugaku Zasshi. 125, 327–334 [DOI] [PubMed] [Google Scholar]
- 45. Tennant J. R. (1964) Evaluation of the trypan blue technique for determination of cell viability. Transplantation 2, 685–694 [DOI] [PubMed] [Google Scholar]
- 46. Ishiyama M., Miyazono Y., Sasamoto K., Ohkura Y., Ueno K. (1997) A highly water-soluble disulfonated tetrazolium salt as a chromogenic indicator for NADH as well as cell viability. Talanta 44, 1299–1305 [DOI] [PubMed] [Google Scholar]
- 47. Pepper C., Thomas A., Tucker H., Hoy T., Bentley P. (1998) Flow cytometric assessment of three different methods for the measurement of in vivo apoptosis. Leuk. Res. 22, 439–444 [DOI] [PubMed] [Google Scholar]
- 48. Naismith J. H., Field R. A. (1996) Structural basis of trimannoside recognition by concanavalin A. J. Biol. Chem. 271, 972–976 [DOI] [PubMed] [Google Scholar]
- 49. Transue T. R., Smith A. K., Mo H., Goldstein I. J., Saper M. A. (1997) Structure of benzyl T-antigen disaccharide bound to Amaranthus caudatus agglutinin. Nat. Struct. Biol. 4, 779–783 [DOI] [PubMed] [Google Scholar]
- 50. Kasai K., Hirabayashi J. (1996) Galectins. A family of animal lectins that decipher glycocodes. J Biochem. 119, 1–8 [DOI] [PubMed] [Google Scholar]
- 51. Weis W. I., Taylor M. E., Drickamer K. (1998) The C-type lectin superfamily in the immune system. Immunol. Rev. 163, 19–34 [DOI] [PubMed] [Google Scholar]
- 52. Vakonakis I., Langenhan T., Prömel S., Russ A., Campbell I. D. (2008) Solution structure and sugar-binding mechanism of mouse latrophilin-1 RBL. A 7TM receptor-attached lectin-like domain. Structure 16, 944–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ogawa T., Watanabe M., Naganuma T., Muramoto K. (2011) Diversified carbohydrate-binding lectins from marine resources. J. Amino Acids 2011, 838914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kotake T., Dina S., Konishi T., Kaneko S., Igarashi K., Samejima M., Watanabe Y., Kimura K., Tsumuraya Y. (2005) Molecular cloning of a β-galactosidase from radish that specifically hydrolyzes {β}-(1–3)-and {β}-(1–6)-galactosyl residues of Arabidnogalactan protein. Plant Physiol. 138, 1563–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stepan H., Pabst M., Altmann F., Geyer H., Geyer R., Staudacher E. (2012) O-Glycosylation of snails. Glycoconj. J. 29, 189–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Velkova L., Dolashka P., Lieb B., Dolashki A., Voelter W., Van Beeumen J., Devreese B. (2011) Glycan structures of the structural subunit (HtH1) of Haliotis tuberculata hemocyanin. Glycoconj. J. 28, 385–395 [DOI] [PubMed] [Google Scholar]
- 57. Gutternigg M., Bürgmayr S., Pöltl G., Rudolf J., Staudacher E. (2007) Neutral N-glycan patterns of the gastropods Limax maximus, Cepaea hortensis, Planorbarius corneus, Arianta arbustorum and Achatina fulica. Glycoconj. J. 24, 475–489 [DOI] [PubMed] [Google Scholar]
- 58. Fujii Y., Sugawara S., Araki D., Kawano T., Tatsuta T., Takahashi K., Kawsar S. M., Matsumoto R., Kanaly R. A., Yasumitsu H., Ozeki Y., Hosono M., Miyagi T., Hakomori S.-I., Takayanagi M., Nitta K. (2012) MRP1 expressed on Burkitt lymphoma cells was depleted by catfish egg lectin through Gb3-glycosphinogolipid and enhanced cytotoxic effect of drugs. Protein J. 31, 15–26 [DOI] [PubMed] [Google Scholar]