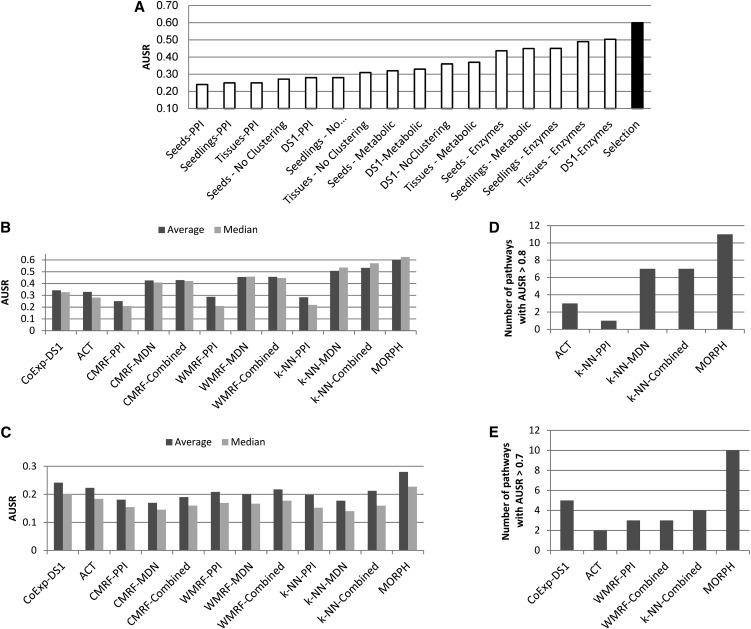
Figure 4.
Performance of the Predictors on 230 Pathways in Arabidopsis.
(A) AUSR scores for different learning configurations. The average AUSR over 66 AraCyc pathways tested is displayed for each combination of gene expression data set and partitioning algorithm (white) and for the selection algorithm (black). Configurations denoted by a data set and PPI or MD network are a combination of a data set and a classifier.
(B) Average and median AUSR scores on 66 AraCyc pathways, using each of the seeds, tissues, and seedlings expression data sets together with PPI and MD networks as input.
(C) Average and median AUSR scores on 164 MapMan pathways, using the same input as in (B).
(D) The number of these pathways that had AUSR score above 0.8 when using MORPH, ACT, and the best performers among the other classifiers in (B).
(E) The number of those pathways that had AUSR score above 0.7 when using MORPH, ACT, and the best performers among the other classifiers in (D). CMRF ranks candidate genes by the number of pathway genes in their neighborhood. WMRF ranks candidates by their similarity with neighbor pathway genes. PPI, methods using the PPI network; MDN, methods using the MD network; Combined, methods using both networks.