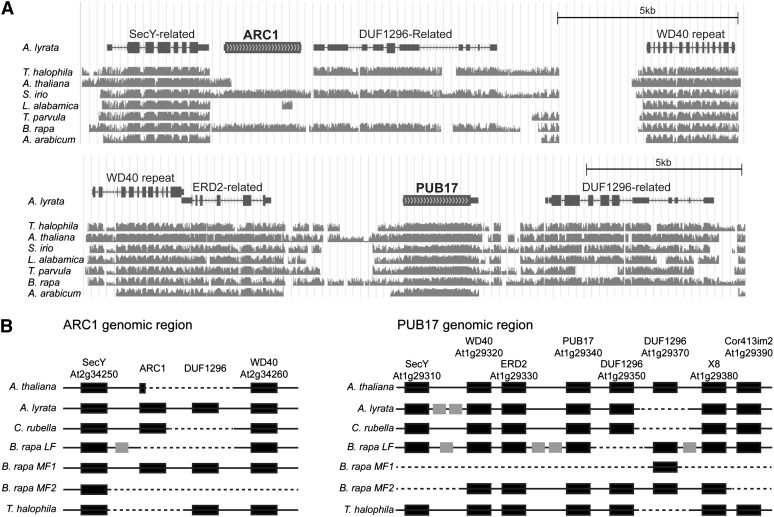
Figure 1.
Synteny of the ARC1 and PUB17 Genomic Regions in the Brassicaceae.
(A) Synteny in the predicted A. lyrata ARC1 and PUB17 genomic regions to T. halophila, A. thaliana, S. irio, L. alabamica, T. parvula, B. rapa, and A. arabicum. This image was generated in the University of California, Santa Cruz genome browser (Fujita et al., 2011) using a nine-species whole-genome orthologous alignment assembled by the VEGI consortium (http://grandiflora.eeb.utoronto.ca:8086). The A. lyrata annotated genes are shown at the top, and the degrees of similarity to the A. lyrata ARC1 or PUB17 genomic regions are shown in gray for each genome. Missing regions relative to the A. lyrata ARC1 or PUB17 genomic regions are shown as gaps.
(B) Schematic illustrating the gene conservation in the ARC1 and PUB17 genomic regions for A. thaliana, A. lyrata, C. rubella, B. rapa (LF, MF1, and MF2 subgenomes), and T. halophila. Annotated Brassicaceae genomes in the public genome databases were used to identify predicted genes present in the ARC1 and PUB17 genomic regions. The A. thaliana annotated genes are shown at the top, and the black boxes represent conserved orthologs annotated in the respective genomes. Gray boxes represent unconserved genes, and dashed lines represent deletions in the scaffolds.