Fig. 1. Identification of a DimA-interacting protein.
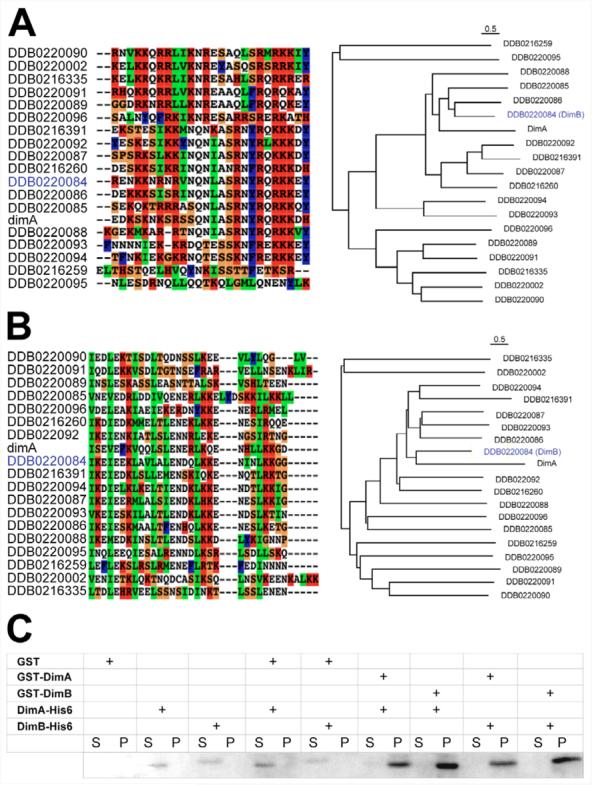
The Dictyostelium genome contains 19 potential bZIP genes. Alignments and phylogenetic trees of the DimA basic DNA-binding domain (A) and leucine zipper dimerisation domain (B) are shown. Identifiers from the Dictyostelium database (www.dictybase.org) are shown with DimB highlighted in blue. Basic residues are shown in red, hydrophobic in blue, small and polar in white, and secondary structure breakers in orange. The basic regions were defined as the first 26 amino acids from the basic DNA-binding domain, and all 19 sequences were aligned using CLUSTALX (default setting) and a phylogenetic neighbour-joining tree was plotted using NJ-Plot. Scale bars above the alignment indicate 0.5 estimated amino acid substitutions per site. Multiple sequence alignment and phylogenetic analysis were performed based on the dimerization domain, represented by the first four hepatads from the leucine zipper region of each protein. The results indicated a greater sequence identity between DimA and DimB bZIP domain. (C) DimA and DimB form homo- and heterodimers in vitro. Fusion proteins were expressed in E. coli and purified by affinity chromatography. DimA or DimB GST fusion proteins were mixed with His tagged versions before incubating in the presence of glutathione sepharose beads. Presence of His fusion proteins was monitored in the supernatant (S, unbound) and pellet (P, bound).
