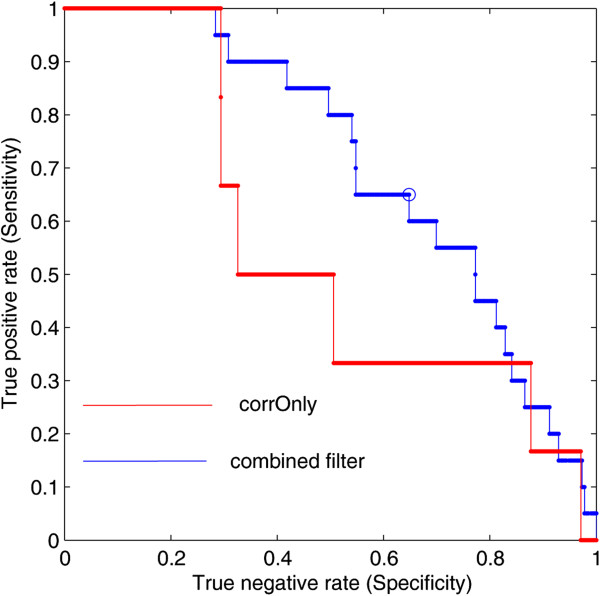
Figure 4.
The ROC curve was created by plotting the sensitivity against specificity. Sensitivity is the fraction of number of true positive assessment versus that of all positive assessment (Sensitivity = TP/ (TP + FN)). Specificity is the fraction of number of true negative assessment versus that of all negative assessment (Specificity = TN/ (TN + FP)). The red line “corrOnly” represents the traditional method to identify CRGs only based on the correlation between gene expression and drug activity. The blue line “combined filter” represents the proposed method to identify CRGs by integrating information from CCRG enriched GO terms and network features of PPIN. The ROC curve was used to evaluate the performance of both methods. For the proposed method, we rank all the genes in HPRD protein interaction network by Q statistics (see details in the Methods section of the manuscript). According to Q statistics and whether the genes are CCRGs, we plotted the ROC curves for our method. While for the traditional correlation method, we ranked all drug-CRG pairs using absolute PCC. According to PCC and whether genes were CCRGs, we also plotted the ROC curves.