Abstract
DNA repair is an essential cellular process required to maintain genomic stability. Every cell is subjected to thousands of DNA lesions daily under normal physiological conditions. Ionizing radiation (IR) is a major DNA damaging agent that can be produced by both natural and man-made sources. A common source of radiation exposure is through its use in medical diagnostics or treatments such as for cancer radiotherapy where relatively high doses are received by patients. To understand the detailed DNA repair gene transcription response to high dose IR, gene expression exon array studies have been performed and the response to radiation in two divergent cell types, lymphoblastoid cell lines and primary fibroblasts, has been examined. These exon arrays detect expression levels across the entire gene, and have the advantage of high sensitivity and the ability to identify alternative transcripts. We found a selection of DNA repair genes, including some not previously reported, that are modulated in response to radiation. Detailed dose and time course kinetics of DNA repair transcription was conducted and results have been validated utilizing PCR methods. Alternative transcription products in response to IR were identified in several DNA repair genes including RRM2B and XPC where alternative initiation sites were found. These investigations have advanced the knowledge about the transcriptional response of DNA repair.
Introduction
Humans are exposed to IR from a number of sources. One of the most common sources of exposure is through medical procedures. For example, IR is commonly used for imaging and cancer treatment. Relatively large doses, on the order of tens of Gray (Gy), are received during radiotherapy that is provided as a curative or palliative treatment for a large proportion of cancer patients. Other sources of IR exposure include natural background radiation, accidental and occupational exposures. IR can cause direct or indirect damage to DNA which can lead to double-strand breaks (dsbs), single-strand breaks, base damage and DNA-DNA and DNA-protein cross-links. DNA dsbs are a critical lesion since they can result in senescence, death, or if improperly repaired, potentially tumorigenesis [1]. However, DNA dsbs occur relatively infrequently compared to other types of DNA damage. Organisms have evolved complex and sometimes redundant ways to repair a variety of DNA damage using a number of different pathways. Many proteins are involved in the pathways which include mismatch repair (MMR), base excision repair (BER), nucleotide excision repair (NER), non-homologous end joining (NHEJ) and homologous recombination (HR).
The importance of these DNA repair genes is evident from consequential disease associated with a particular DNA repair gene dysfunction. Commonly, DNA repair deficient individuals show sensitivity to DNA damaging agents and are susceptible to cancer [2], [3]. Gene knockout studies have shown some DNA repair genes are critical for an organism’s survival since they can result in embryonic lethality [4], [5]. Deficiencies of many genes in DNA repair pathways have been characterized and often result in clinical pathologies. Mutations in DNA dsb repair genes, including LigIV, and DNAPKcs of the NHEJ pathway have been identified in humans and result in radiosensitivity [6], [7]. Hereditary non-polyposis colorectal cancer (HNPCC) can be due to defects in genes that are required for mis-match DNA repair pathway, such as MSH2, HLH1 or MSH6 [8], [9]. A number of diseases are the result of a defect in NER. These diseases include xeroderma pigmentosum (XP) which is due to a defect in one of approximately eleven XP associated genes (including XPA, ERCC3 (XPB), XPC, and POLH (XPV)). Skin cancer commonly occurs in these patients who are unable to properly repair UV induced DNA damage [10]. Trichothiodystrophy (mutations in ERCC2 and ERCC3) and Cockayne syndrome (includes mutations in ERCC6 and ERCC8 genes) are other NER-associated diseases with similarities to XP but characterized by slightly different phenotypes [11], [12]. Fanconi anemia is another DNA repair deficiency disease. There are at least 13 DNA repair genes that are associated with this disease for which affected patients are especially sensitive to inter-strand DNA cross-linking [13]. There are approximately 153 genes that are directly involved with DNA repair [14], [15]. It should be noted that there are many other additional factors which are associated with these DNA repair proteins, or contribute to the proper regulation of DNA damage repair.
Most DNA repair occurs relatively rapidly following DNA damage. Sensors and transducer proteins organize the effector proteins for repair. Very rapid DNA repair responses often occur through post-translational protein modifications. For example, phosphorylation [16] by PI3 protein kinases such as ATM, ATR and DNA-PK is a well known regulatory mechanism. However, DNA repair gene transcription is also modulated in response to DNA damage [17], [18], [19], [20], [21], [22]. Transcription has many levels of regulation ranging from initiation to processing and transport of the mature message. High density gene expression arrays containing probes for every exon offer a means to conduct a detailed survey of the whole transcriptome at the exon level which can reveal alternative transcription following DNA damage.
DNA repair dysfunction commonly leads to disease including cancer, therefore, understanding the molecular mechanisms of DNA repair genes in response to DNA damaging agents such as IR is critical to develop innovative treatments. Therefore, we have comprehensively assessed the transcription of the known DNA repair genes [14] in response to IR exposure utilizing an exon array platform where each gene is extensively covered by probes, thus yielding highly robust data and the ability to assess alternative transcription.
Materials and Methods
Cell Culture
Derivation of lymphoblast and primary fibroblast cell lines has been previously described [19], [23]. Transcriptional response using exon arrays for twelve individuals were analysed [19], [23], [24] at 0 and 10 Gy 4 hr post-IR for both Epstein Barr virus (EBV) transformed lymphoblastoid cell lines (LCLs) and primary fibroblasts. Four samples were used from each group for other time points and doses and two samples of unirradiated cells for baseline normalization. LCLs were grown in RPMI media supplemented with 10% FBS and gentamicin and incubated in a 5 percent CO2 humidified 37°C incubator. All patients have given written informed consent and studies have been approved by the Peter MacCallum Cancer Centre and Monash University ethics committee.
RNA Isolation
Cells (1×107) were pelleted and resuspended in 3 ml PBS. Equal volume of Trizol (Invitrogen, Carlsbad, CA, USA) was added, mixed and incubated at room temperature for 15 minutes followed by the addition of 200 µl of chloroform. The sample was centrifuged, the aqueous layer was collected and mixed with and equal volume of 70 percent ethanol and added onto a RNeasy column (Qiagen, Venlo, The Netherlands). The RNA extraction was continued by using the RNAeasy method (with DNAse treatment) as per manufacturer’s recommendation except starting with the addition of Buffer RW1 to the sample. RNA concentration and integrity was determined by analysing on a bioanalyzer (Agilent, Santa Clara, CA, USA). RNA was determined to be high enough quality if a minimum RIN of 8.5 was obtained.
Exon Arrays
GeneChip Human Exon 1.0 ST Array analysis was performed as per the ‘GeneChip Whole Transcript (WT) Sense Target labelling assay Manual’ (Affymetrix, Santa Clara, CA, USA). The rRNA from 1 µg of total RNA was reduced using a RiboMinus Human/Mouse Transcriptome Isolation Kit (Invitrogen, Carlsbad, CA, USA). Assessment of array quality was determined using Expression Console (Affymetrix.com). Gene expression was assessed using R, normalized using RMA and analysed using significance analysis of microarrays (SAM [25]). Probe selection regions (PSRs) are regions of primer sets designated for exons or potential exons in particular genes. Note that PSR probe sets have probe specific fluorescent and therefore show different relative levels of fluorescence. Array data and normalized expression have been deposited in the gene expression omnibus database: accession number GSE41840.
Transcriptional Validation
Primers were designed to candidate exons or genes using ‘primer 3′ or Primer-Blast on-line software (NCBI). cDNA was made from 1 µg RNA using Super Script III First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, USA) as per manufactures recommendation. Initially the RNA, dNTPs and random hexamers were heated to 65°C for 5 minutes and then incubated at 25°C for 5 minutes with a subsequent incubation of 50°C for 1 hour and a 70°C incubation for 15 minutes with first strand buffer (Invitrogen, Carlsbad, USA), 0.1 M DTT, 0.5 mM dNTPs, 250 ng of random hexamers, 40 units of RNaseOUT and 200 units of SuperScript III RT. PCR amplification was carried out using 1.25 Units Go Taq polymerase (Promega, Wisconsin, USA), 200 nM primers, 5 ng cDNA, with a cycling protocol of 95°C: 2 min; (95°C: 15 sec; 60°C: 45 sec; 72°C: 30 sec) ×30; 72°C: 5 min. Products were run on a 2 percent agarose gel to determine amplification of the proper sized product. Real-time PCR was performed using these primers under the following conditions: Sybr Green Master Mix (Applied Biosystems, United Kingdom) with 200 nM of each primer was mixed with 5 ng of cDNA. The cycling steps were as follows. 95°C: 10 min; (95°C: 15 sec; 60°C: 60 sec) ×40, with a melting temperature ramp following amplification. A robotic system was used to load a 384 well plate with a subsequent run on the ABI 7900 quantitative real time PCR machine. All samples were run in triplicate. Primers used are shown in Table S1.
5′ RNA Ligase-mediated Rapid Amplification of cDNA Ends (5′-RLM-RACE)
5′-RLM-RACE was performed using FirstChoice RLM-RACE kit (Ambion, Austin, TX, USA) recommended by the manufacturer except the CIP digested RNA was purified using RNAeasy kit (Qiagen, Venlo, The Netherlands). The RRM2B and XPC transcripts were amplified using semi-nested PCR (as recommended by Ambion) with forward (inner and outer) primers to the adaptor (provided with the FirstChoice RLM-RACE kit) and reverse gene-specific primers (Table S1).
Sequencing of PCR Amplicons
PCR amplicons were separated on 2 percent agarose, bands were stabbed using a pipet tip, placed in 100 µl of water, and 1 µl was re-amplified. The re-amplified PCR product was cleaned-up using a Qiagen PCR product spin column. Big Dye terminator sequencing was performed using PCR primers and the transcription start sites at the nucleotide level were determined by sequence comparison (NCBI BLAST).
Results
DNA Repair Genes Modulated at the Transcription Level
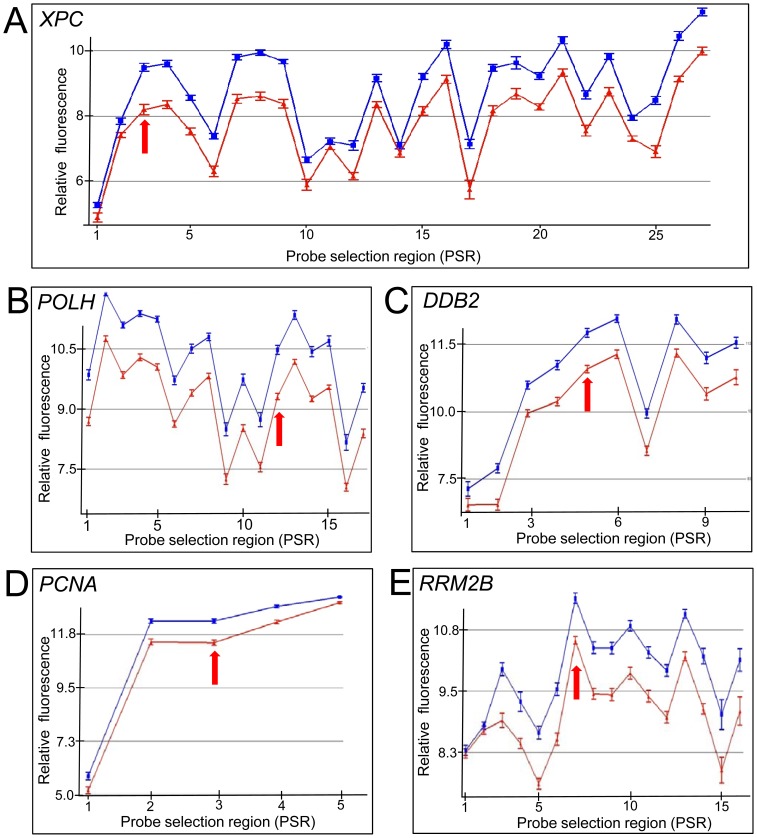
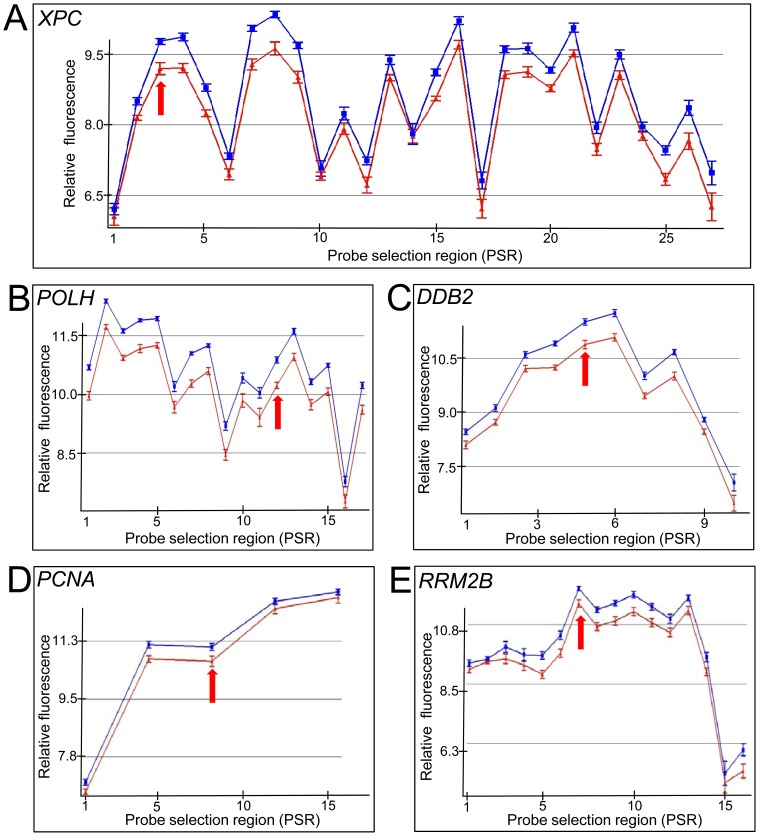
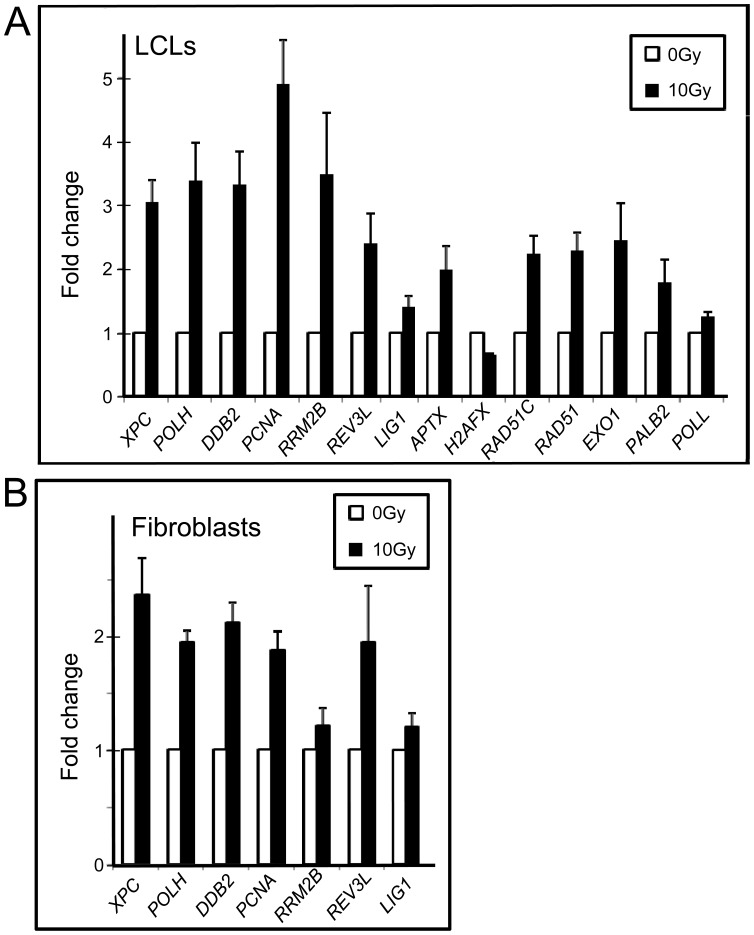
The effect of IR on DNA repair gene expression in cells derived from two different lineages, LCLs and primary fibroblasts, was determined. Exon arrays were used to provide comprehensive probe coverage for all known DNA repair gene exons [14], [15]. The DNA repair genes that showed modulation were ranked based on ANOVA p-values (Tables 1 and 2). We found that 21 DNA repair genes were modulated in LCLs and 16 in primary fibroblasts, respectively when using a p-value cut-off of <0.05 comparing sham-irradiated to those irradiated with 10 Gy at 4 hrs post-IR. The top five genes (XPC, POLH, DDB2, PCNA and RRM2B) in lymphoblastoid cells, which were also induced in fibroblast cells, showed a clear gene expression induction at most exons across the gene (Figures 1 and 2, Profiles of all genes listed in the tables with probe selection regions noted are shown in Figures S1 and S2). These five genes have previously been identified as responsive to DNA damaging agents including IR [17], [19], [26], [27], [28], [29], [30], but these have not been well-characterized across the gene at the exon level which reveals underlying features of the transcripts with regard to different isoforms. Some DNA repair genes shown in Tables 1 and 2 (LIG1, PALB2, CHAF1A, and MBD4), which have minor transcriptional changes at 4 hours post-IR, have not previously been recognized to be responsive to IR in this context. We validated the exon array expression data using qRT-PCR for many of the genes listed in Tables 1 and 2 with array data showing a 1.2 fold induction or greater in LCLs at 4 hr post-IR (Figure 3). qRT-PCR values were consistent with the exon array derived data, however, the amount of modulation was augmented when analysed with qRT-PCR. qRT-PCR was also used to determine the response to IR for other DNA repair genes that had p-values derived from the exon array data of >0.05 in LCLs. For example, POLL was shown to have a statistically significant induction (p<0.05) in response to IR in LCLs (Figure 3).
Table 1. DNA repair genes transcriptionally modulated in LCLs at 4 hr post-IR (p<0.05).
| Gene | Rank | Total gene rank | Fold change | p-value | DNA repair pathway |
| XPC | 1 | 17 | 2 | 1.4E−10 | Nucleotide excision repair |
| POLH | 2 | 42 | 2.2 | 8.2E−09 | DNA polymerases (catalytic subunits) |
| DDB2 | 3 | 48 | 1.7 | 2.3E−08 | Nucleotide excision repair |
| PCNA | 4 | 54 | 1.5 | 4.6E−08 | DNA polymerases (catalytic subunits) |
| RRM2B | 5 | 69 | 1.8 | 2.1E−07 | Modulation of nucleotide pools |
| REV3L | 6 | 204 | 1.3 | 0.0001 | DNA polymerases (catalytic subunits) |
| LIG1 | 7 | 298 | 1.2 | 0.0005 | Nucleotide excision repair |
| APTX | 8 | 383 | 1.2 | 0.001 | Editing and processing nucleases |
| TDP1 | 9 | 411 | −1.2 | 0.002 | Repair of DNA-protein crosslinks |
| H2AFX | 10 | 512 | −1.2 | 0.003 | Chromatin Structure |
| CHAF1A | 11 | 538 | 1.1 | 0.004 | Chromatin Structure |
| MBD4 | 12 | 638 | −1.2 | 0.006 | Base excision repair |
| ATM | 13 | 672 | −1.2 | 0.007 | Genes defective in diseases associated with sensitivity to DNA damaging agents |
| DCLRE1C | 14 | 691 | −1.2 | 0.007 | Non-homologous end-joining |
| RAD51C | 15 | 696 | 1.4 | 0.007 | Homologous recombination |
| RAD51 | 16 | 729 | 1.2 | 0.009 | Homologous recombination |
| EXO1 | 17 | 850 | 1.2 | 0.013 | Editing and processing nucleases |
| RAD51L3 | 18 | 946 | 1.1 | 0.018 | Homologous recombination |
| PALB2 | 19 | 963 | 1.1 | 0.018 | Genes defective in diseases associated with sensitivity to DNA damaging agents |
| MDC1 | 20 | 1179 | −1.1 | 0.030 | Other conserved DNA damage response genes |
| RECQL4 | 21 | 1403 | 1.1 | 0.042 | Genes defective in diseases associated with sensitivity to DNA damaging agents |
Table 2. DNA repair genes transcriptionally modulated in primary fibroblasts at 4 hr post-IR (p<0.05).
| Gene | Rank | Total gene rank | Fold change | p-value | DNA repair pathway |
| POLH | 1 | 12 | 1.6 | 4.1E−07 | DNA polymerases (catalytic subunits) |
| DDB2 | 2 | 15 | 1.5 | 9.7E−07 | Nucleotide excision repair |
| XPC | 3 | 19 | 1.4 | 1.3E−06 | Nucleotide excision repair |
| RRM2B | 4 | 172 | 1.4 | 0.003 | Modulation of nucleotide pools |
| DCLRE1C | 5 | 272 | −1.2 | 0.008 | Non-homologous end-joining |
| TDP1 | 6 | 375 | −1.2 | 0.014 | DNA protein crosslinks |
| DCLRE1A | 7 | 449 | −1.2 | 0.018 | DNA cross-link repair |
| CHEK2 | 8 | 485 | −1.1 | 0.020 | Effector kinase |
| ALKBH2 | 9 | 509 | −1.1 | 0.022 | Resistance to alkylation damage |
| PCNA | 10 | 570 | 1.3 | 0.026 | DNA polymerases (catalytic subunits) |
| DUT | 11 | 592 | −1.1 | 0.027 | dUTPase |
| UBE2A | 12 | 739 | −1.2 | 0.035 | Editing & processing nuclease |
| XAB2 | 13 | 751 | 1.1 | 0.036 | Nucleotide excision repair |
| MNAT1 | 14 | 888 | −1.1 | 0.044 | Nucleotide excision repair |
| NEIL3 | 15 | 997 | −1.4 | 0.049 | Base excision repair |
| APEX1 | 16 | 998 | −1.1 | 0.049 | Base excision repair |
Figure 1. Induction of DNA repair genes at the exon level four hours after treatment with 10 Gy IR in LCLs.
The top five DNA repair genes: XPC (A), POLH (B), DDB2 (C), PCNA (D) and RRM2B (E) as identified using Partek Genomics Suite 6.6 statistical package. Relative fluorescence (y-axis; log2) is plotted for each PSR (x-axis). Core PSRs are labelled numerically in a 5′ to 3′ direction (left to right). Samples were either sham irradiated (red) or irradiated (blue) with 10 Gy from a 137Cs source. Arrow indicates the PSR region to which primers were designed for qRT-PCR used in figure 3A. Error bars = SEM (n = 12).
Figure 2. Induction of DNA repair genes at the exon level four hours after treatment with 10 Gy IR in primary fibroblast cells.
The DNA repair genes: XPC (A), POLH (B), DDB2 (C), PCNA (D) and RRM2B (E) as identified using Partek Genomics Suite 6.6 statistical package. Relative fluorescence (y-axis; log2) is plotted for each PSR (x-axis). Core PSRs are labelled numerically in a 5′ to 3′ direction (left to right). Samples were either sham irradiated (red) or irradiated (blue) with 10 Gy of radiation from a 137Cs source. RNA was collected 4 hours following treatment. Arrow indicates the PSR region to which primers were designed for qRT-PCR used in figure 3B. Error bars = SEM (n = 12).
Figure 3. Validation of DNA repair gene expression modulation following 10 Gy IR using qRT-PCR.
Ct values were normalized to PGK. Each bar represents data from 12 different cell lines for both LCL (A) and primary fibroblasts (B) with the following exceptions: 6 samples were used for PCNA and RRM2B in the LCL experiments; 10 samples were used for XPC, RRM2B, REV3L for the fibroblast experiments and 5 samples for, EXO1, PALB2, LIG1 and H2AFX were used in the fibroblast experiments. Gene expression levels were averaged across multiple experiments. Four separate qRT-PCR runs were carried out for POLH, DDB2, APTX, RAD51C, and PALB2 genes; three separate qRT-PCR runs were carried out for PCNA, REV3L and EXO1 genes; and two separate qRT-PCR runs were carried out for XPC, RRM2B, H2AFX, and RAD51 genes. Error bars = SEM (n = 12). Each value on an experiment was run in triplicate. All differences shown are statistically significant (p<0.05) using a t-test. PSRs used for amplification are: XPC: PSR853; POLH: PSR124; DDB2: PSR663; PCNA: PSR213; RRM2B: PSR293; REV3L: PSR729; LIG1: PSR905; APTX: PSR338; H2AFX: PSR185; RAD51C: PSR786; RAD51: PSR100; EXO1: PSR239; PALB2: PSR346; POLL: PSR904 for which some are indicated by and arrow in figures 1 and 2 and all full PSR numbers can be found in Figures S1 and S2.
DNA Repair Gene Expression in Two Divergent Cell Types
The radiation response between the two cell types were compared and found to show a similar response for most DNA repair genes, however, the LCL response generally had a larger induction at the 4 hour time point compared to the fibroblast cell lines. This is especially evident for PCNA and RRM2B genes, where there is more than a two-fold difference between the cell types (Figures 1,2,3; Tables 1 and 2). Dose response and time course experiments were consistent with this observation (Figures 4,5,6). However, microarray expression levels (Figures 1 and 2) and additional qRT-PCR data (Figure S3) indicate that the initial levels of transcripts for XPC and RRM2B are higher in the fibroblasts compared to the LCLs (relative to the control gene PGK).
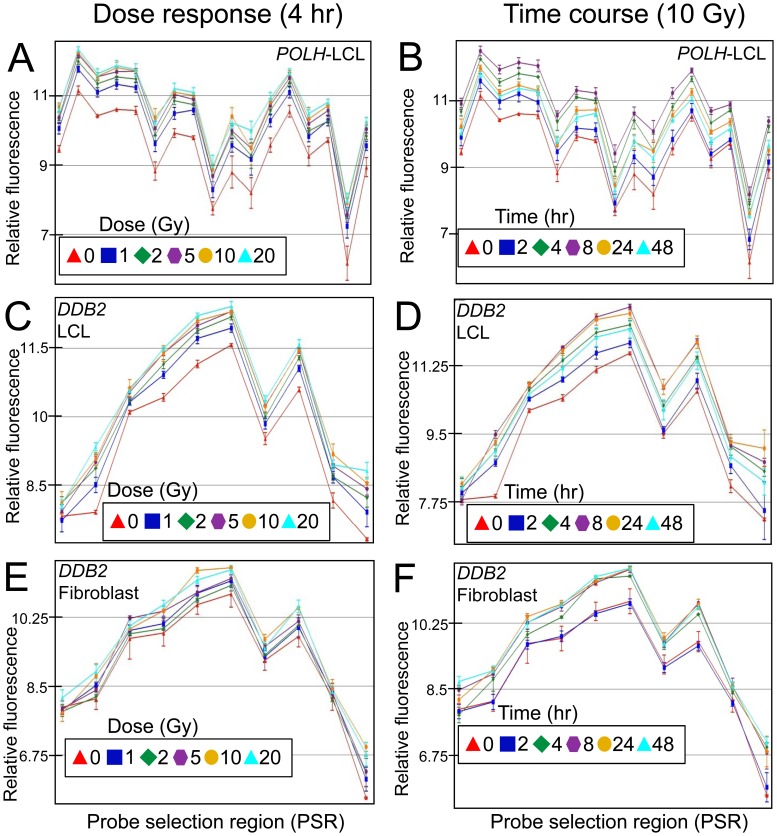
Figure 4. Dose response and time course of selected DNA repair genes in human cell lines.
PSR hybridization signals are shown for a two DNA repair genes that are induced following radiation. These are POLH (A, B) and DDB2 (C–F) in LCLs (A–D) or fibroblasts (E, F). Relative fluorescence is plotted on the y-axis and PSRs are plotted evenly across the x-axis in a 5′ to 3′ direction (left to right). Samples from different individuals were either sham irradiated (red) or irradiated with 1 Gy (blue), 2 Gy (green), 5 Gy (purple), 10 Gy (orange) or 20 Gy (aqua) of radiation (A, C and E; n = 4). RNA was collect 4 hours following treatment. Time course of DNA repair genes were either sham irradiated (red) or irradiated with 10 Gy and RNA isolated 2 hrs (blue), 4 hrs (green), 8 hrs (purple), 24 hours (orange) or 48 hours (aqua) after irradiation (B, D and F; n = 4). Error bars = SEM.
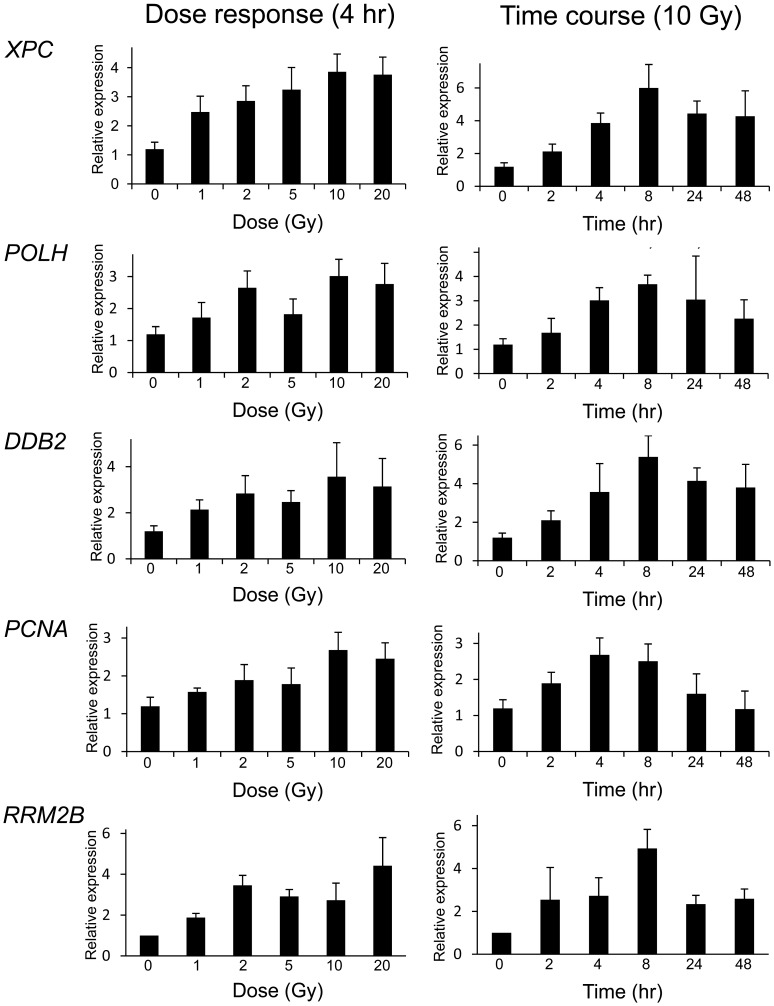
Figure 5. Expression levels of the top five DNA repair genes as determined by qRT-PCR in LCLs over different IR doses and times.
Relative gene expression is shown for samples that were either sham irradiated or irradiated with 1 Gy, 2 Gy, 5 Gy, 10 Gy or 20 Gy of radiation at 4 hours post-IR, or sham irradiated or irradiated with 10 Gy at 2 h, 4 h, 8 h, 24 h or 48 h post-IR.
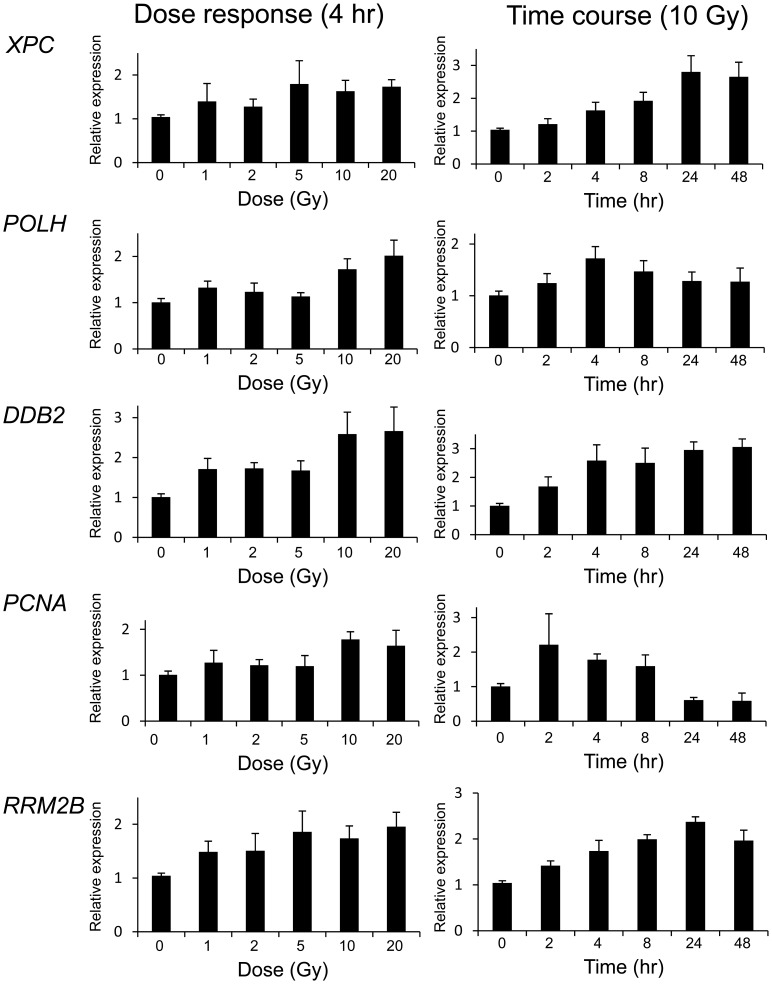
Figure 6. Expression levels of the top five DNA repair genes as determined by qRT-PCR in fibroblasts over different IR doses and times.
Relative gene expression is shown for samples that were either sham irradiated or irradiated with 1 Gy, 2 Gy, 5 Gy, 10 Gy or 20 Gy of radiation at 4 hours post-IR, or sham irradiated or irradiated with 10 Gy at 2 h, 4 h, 8 h, 24 h or 48 h post-IR.
DNA Repair Gene Expression dose Response and Time Course
The effect of dose on DNA repair gene expression response was investigated using whole genome exon arrays at four hours post-treatment using both LCLs and primary fibroblasts (Figure 4). Some genes such as POLH, had a robust response to a relatively low dose of 1 Gy, and then consistent but smaller incremental responses to IR with increasing doses of radiation up to 20 Gy (Figure 4A). DDB2 showed a similar pattern to POLH in LCLs but the fibroblast group showed a more gradual transcript level increase with dose (Figures 4C and 4E). These results were consistent with qRT-PCR expression analysis (Figures 5 and 6). Other genes such as XPC, PCNA and RRM2B showed a dose dependent response in both LCLs and fibroblasts although these responses were generally less in fibroblasts (Figures 5 and 6). An additional set of 12 arrays of the fibroblast samples isolated 4 hr after 2 Gy IR were also performed, and although the inductions were generally less, the results were consistent with the 10 Gy responses in this cell type (data not shown).
DNA repair gene expression across different time points was investigated up to 48 hrs post-irradiation in both LCLs and fibroblasts treated with 10 Gy. Early robust responses were identified 2 hours post-IR in LCLs for POLH and DDB2 showing peak expression 8 hours after treatment, followed by a decrease after 24 hours, and the levels of transcripts were still above basal expression levels at 48 hours (Figure 4). Temporal patterns of gene modulation were validated using qRT-PCR for these genes and for XPC, PCNA and RRM2B (Figure 5). The results were consistent between exon array and qRT-PCR data. The peak expression levels in the fibroblast cells were different from the LCLs for XPC and RRM2B. In the LCLs, expression levels were highest at 8 hours post-irradiation and decreased by 48 hours whereas in the fibroblast cells the expression did not peak until 24 hours and was still elevated at 48 hours (Figures 5 and 6). The DDB2 expression levels in the fibroblast cells differed slightly to the levels in LCLs. The expression levels in both cell types peaked at 8 hours. However, in the LCLs, the levels start dropping 24 hours post irradiation but they remain high in the fibroblasts up to 48 hours post irradiation (Figures 3, 4, 5 and 6). For POLH, and PCNA, peak expression levels were similar in the LCLs and fibroblasts (Figures 5 and 6).
Alternative Transcription in DNA Repair Genes
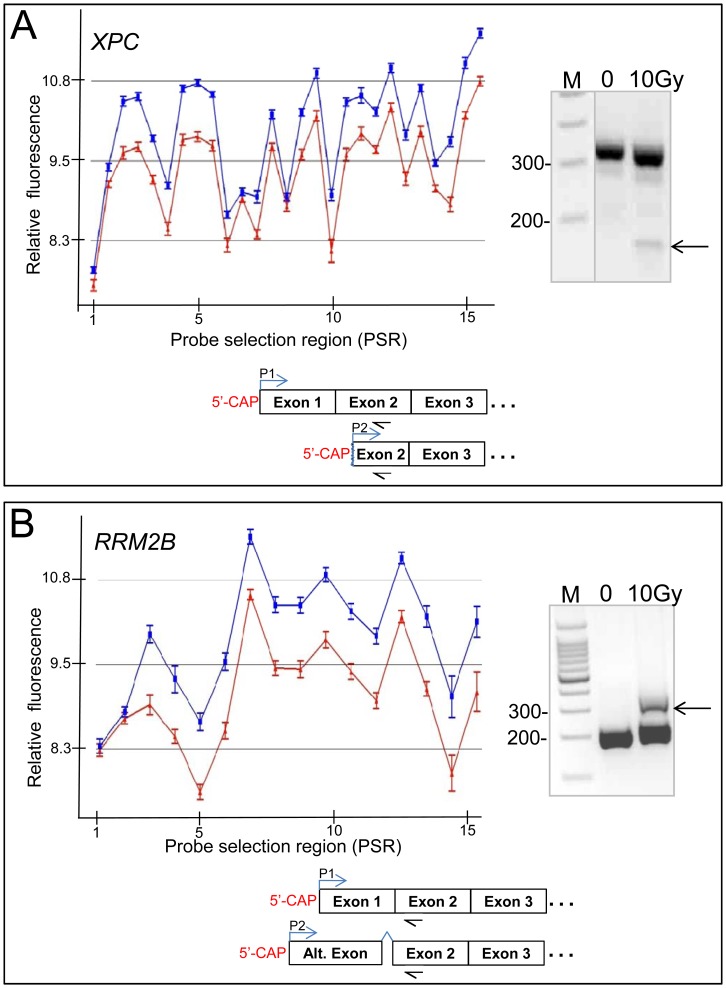
The comprehensive exon coverage of this array platform enabled us to investigate alternative transcription products. Certain DNA repair genes showed variations across the gene in that some PSRs showed different levels of expression changes after treatment with IR. This suggests induction or changes in levels of alternative transcripts following irradiation. For example, PCNA showed features of alternative termination (Figure 1D). Other genes, such as XPC and RRM2B showed a robust level of expression in all but the first two 5′ PSRs (Figure 7). To confirm the use of an alternative transcriptional initiation sites we performed 5′-RLM-RACE using RNA transcripts from LCLs for XPC and RRM2B. A shorter XPC transcript was identified after IR. Sequencing (Figure S4) of PCR amplified amplicon (150 bp; Figure 7A) confirmed that the start site of this shorter XPC transcript occurred within exon two, 261 bp 3′ of the full length transcript’s initiation site (NCBI sequence NM_004628). This transcript is similar to the NCBI XPC sequence X65024 which has a transcription start site 265 bp 3′ of the full length transcript. The predicted isoform (NCBI protein id: CAA46158.1) encodes for a protein missing 117 amino acids from the N-terminus of the full length XPC protein isoform. The 5′-RLM-RACE PCR data suggest this radiation induced shorter transcript is probably a minor product (Figure 7A) and the full length transcript is also radiation induced. Prediction of Protein Sorting Signals and Localization Sites in Amino Acid Sequences (PSORT) analysis (http://psort.hgc.jp) of the predicted proteins suggests that the missing N-terminus region contained several coiled-coil regions (Lupas’s algorithm).
Figure 7. Alternative transcripts in DNA repair genes are induced by IR.
Alternative transcripts in the DNA repair genes, XPC (A) and RRM2B (B) in response to IR are shown. Graph axes are as in figure 1; PCR products from 5′-RML-RACE were run on a 2% agarose gel. An arrow indicates the amplicon from the alternative initiated transcript that was sequenced (gel picture to the right of the panel; Figure S4). Diagrams of the predominant transcripts (initiating by P1) and the alternatively initiated (P2) transcripts after IR are shown below. Primer locations for 5′ RLM-RACE are indicated below exon 2 (arrow pointing to the left).
A longer alternative transcript was identified in RRM2B after irradiation as depicted by a 334 bp PCR amplicon (Figure 7B) which was identified by sequencing (Figure S4) to be a transcript with a start site at nucleotide 35 of the published variant 2 transcript sequence (NCBI: NM_001172477). The predicted isoform 2 would be missing the first 16 amino acids of the predominant isoform 1 protein and would have an additional 88 unique amino acids at the N-terminus (NCBI protein NP 001165948). The exon array expression profile from the fibroblasts also supports the expression of this longer variant 2 transcript after IR (Figure 1E). PSORT analysis (κ-NN prediction) predicts that the isoform 1 has a 61 percent chance to be directed towards cytoplasmic localization with a 26 percent chance for nuclear localization and a 9 percent chance for mitochondrial localization. However, isoform 2 is predicted to have a 43 percent chance for localization to the mitochondria, with only a 35 and 22 percent chance for cytoplasmic or nuclear localization, respectively.
Discussion
These investigations have provided a comprehensive investigation of the transcription response of DNA repair genes to DNA damage caused by IR on the exon level in two divergent cell lineages. Little is known about alternative transcripts which are modulated by IR and this aspect of transcription has been addressed. Thirty DNA repair genes were identified as radiation responsive at the transcript level. Many of these DNA repair genes did have relatively low levels of transcriptional modulation at the early time point of 4 hours. Some of these genes have not previously reported to be responsive to IR. The small changes for some of these transcripts may be why other less sensitive analyses failed to detect them. Some genes showed modest lower expression levels after IR. The products of the down-regulated genes may be required for cell cycle progression and replication where proofreading and other DNA repair activity is needed during these normal cell processes, and may be a reflection of the IR induced cell cycle arrest. IR produces free radicals resulting in oxidative stress [31]. Repair of oxidative DNA lesions involve the BER and NER DNA repair pathways [32]. Our study has shown several NER pathway genes (XPC, DDB2, and LIG1) and a BER pathway gene (MBD4) are modulated by IR (Tables 1 and 2).
Effects of dose and time after IR on the DNA repair gene transcription was also investigated to gain further insights into the kinetics of the IR-induced DNA repair gene transcriptional response. The dose kinetics of the DNA repair genes in most cases showed increased gene expression at the higher doses, but commonly reached a maximal level earlier than the maximal dose indicating a plateau effect. Many of the genes also showed a distinct maximal response prior to 48 hours post-IR. The variation in response kinetics can reveal some insight into the responses such as those genes that may be coordinately regulated in response to p53 such as XPC, RRM2B, DDB2 and POLH.
Alternative transcripts of RRM2B and XPC in response to damage were also identified. In both these genes, the 5′UTR and N-terminus of the predicted protein isoforms differ from that of the predominant transcript. The RRM2B gene product (p53R2) is a p53 regulated ribonuclease reductase small subunit which supplies deoxyribonucleoside triphosphates to sites of DNA damage during DNA repair [33]. p53R2 is known to be induced by DNA damaging agents such as IR and is involved in the p53 regulated cell cycle arrest checkpoint [33]. p53R2 is usually distributed throughout the cytoplasm, but it accumulates in the nucleus following DNA damage induced by UV irradiation [34]. The first 113 amino acids of p53R2 of the most highly expressed transcript (isoform 1) are reported to be critical for the interaction with N-terminus of the cell cycle regulator, p21 [34]. When p53R2 and p21 are located in the nucleus, the interaction between the two proteins decreases and there is a concurrent increase in ribonuclease activity [34]. Although isoform 2 has been reported to the NCBI data system (NCBI: NP 001165948), no reference to its function or specific expression has been described. The p53R2 isoform 2 is missing the first 16 amino acids present in isoform 1 and has an additional 88 unique amino acids at its N-terminus. As the first 113 amino acids of isoform 1 are involved in binding p21, this interaction is probably altered in the IR induced isoform. Another possible aspect of isoform 2 is that it may have altered cellular localization. The radiation responsive gene product, FBXW7, is an example of a protein that has isoforms that differ in their N-terminus which show different subcellular localization [35]. It is possible that the damage induced p53R2 isoform 2 may localize to DNA containing organelles such as the mitochondria and nucleus rather than the cytoplasm. The difference in the N-terminus may also affect the association with p21.
We confirmed that XPC is induced by IR at the transcription level, which has been consistently observed in several studies [18], [30]. XPC is involved in DNA damage sensing for NER. XPC recognizes various forms of DNA damage that result in distortion of the DNA helix resulting in short sections of single stranded DNA [36], [37]. XPC binds to the undamaged nucleotides identifying the template strand for DNA repair, then recruits transcription factor IIH (TFIIH) complex [38]. Recently, XPC has been shown to enhance UV-induced apoptosis by directly binding to the promoter of caspase-2S gene preventing its transcription [39]. The IR induced XPC transcript that we identified using RACE (Figure 7) is a minor product compared to the normally expressed XPC transcript and the resulting predicted protein product, would be missing the first 117 amino acids. XPC protein N-terminus region contains several coiled-coil regions but has not been well characterized. However aa 156–325 have been shown to interact with DNA and XPA [40]. The C-terminus of XPC (aa 492–940) binds to DNA and also interacts with various proteins including RAD23B, CENTN2, and TFIIH. A mRNA transcript (NCBI: X65024) similar to that identified in this study was previously identified in a cDNA clone (XPCC). The cDNA clone increased UV-radiation survival when transfected into a XPC derived cell line [41] indicating that this transcript is capable of producing a functional protein. The functional difference between the two XPC isoforms is presently unknown.
The 5′UTR and N-terminus sequences are known to be responsible for the efficiency of translation, stability, subcellular localisation, cellular and tissue specific expression. It is known that N-terminus sequences often define protein half-life [42]. This may be a crucial mechanism to provide specific function to repair in response to DNA damage and crosstalk with pathways regulating cell cycle and apoptosis. The importance of the induction of alternative transcripts in the DNA damage response has been described for several genes including MDM2 and CDKN1A. DNA damage has been shown to induce isoforms, in a p-53 dependent manner, in both MDM2 [43], [44], [45], [46] and CDKN1A [47]. In the case of MDM2, radiation induces a shorter transcript by using a p53-regulated alternative start site. p53 plays a major role in induction of IR-induced expression of XPC, PCNA, RRM2B, DDB2 and POLH [48], [49], [50], [51] and may also be involved in the induction of the alternative transcripts induced by IR detected in our studies.
The responses to IR between two cell lineages, LCLs and fibroblasts, were compared. Although much of the DNA repair transcription response to IR was similar, it was not identical. The different levels of initial transcript, induction times, and levels after irradiation shown for some specific genes may contribute to the unique cellular responses attributed to these two different cell types. We have found a number of NER repair genes (e.g. XPC, POLH (XPV), DDB2 (XPE) and other non-dsb repair genes (e.g. PCNA, RRM2B) to be induced by IR. The induction of transcription for several NER genes by IR (Table 1) might not only play a role in DNA damage repair but also in a cell fate decision. In fact, studies have shown that two NER proteins, XPC and POLH (XPV), enhance apoptosis [39], [48]. The fact that the expression of a number of NER proteins are up-regulated, and induction of alternative transcripts is observed along with the changes in expression of several apoptosis mediators ([19] and unpublished results) to a higher extent in the radiosensitive LCLs, brings out the potential role for these NER associated proteins in apoptosis. It may have been expected that DNA dsb repair genes such as those found in NHEJ and HR would be induced after IR given their critical nature, but perhaps the numbers of these types of breaks that are normally encountered did not warrant the evolutionary development of regulation of these repair genes at the transcription level in these time frames and regulation at the protein level is a more efficient mechanism for immediate DNA dsb repair. Furthermore, the transcriptional responses for many DNA repair genes may be too subtle for detection.
Conclusion
This investigation provides a comprehensive study of DNA repair gene expression dynamics at the exon level in response to the DNA damaging agent, IR, in human cells. These data provide IR response kinetics covering a range of doses and times, probing at the exon level. Novel expression of alternative transcripts has been examined and DNA repair genes that utilize a mechanism of alternative transcription initiation following IR have been identified.
Supporting Information
Modulation of DNA repair genes at the exon level 4 hours after treatment with 10 Gy IR in lymphoblast cell lines. PSR expression levels are plotted for each of the DNA repair genes with a p-value of <0.05 using Partek Genomics Suite statistical package. Relative fluorescence (y-axis; log base 2) is plotted for each PSR (x-axis). Core PSRs are labelled below the graphs. Samples were either sham irradiated (red) or irradiated (blue) with 10 Gy from a 137Cs source. Error bars = SEM (n = 12).
(PDF)
Modulation of DNA repair genes at the exon level 4 hours after treatment with 10 Gy IR in fibroblast cells. PSR expression levels are plotted for each of the DNA repair genes with a p-value of <0.05 using Partek Genomics Suite statistical package. Relative fluorescence (y-axis; log base 2) is plotted for each PSR (x-axis). Core PSRs are labelled below the graphs. Samples were either sham irradiated (red) or irradiated (blue) with 10 Gy from a 137Cs source. Error bars = SEM (n = 12).
(PDF)
Real-time PCR comparison of DNA repair gene expression between LCLs and fibroblasts. Initial levels of transcripts are significantly higher in fibroblasts (FB) for XPC and RRM2B relative to LCLs, but after IR induction, gene expression levels are not significantly different. Values have been normalized to PGK gene expression. The p-values for differences of initial levels between LCL and fibroblast samples are as follows: XPC: p = 0.0025 and for RRM2B: p = 0.0030.
(PPTX)
Sequence analysis of the 5′RACE amplicons for RRM2B (A) and XPC (B). Sequences homologous to the primers used for their amplifcation are indicated by long horizontal arrows below the sequence. The adaptor sequences are shown in capital letters. The vertical arrows indicate the start site for the capped mRNA transcript. This is nucleotide 35 of NCBI RRM2B sequence NM_001172477 (A), and at nucleotide 261 of NCBI XPC sequence NM_004628 (B).
(PPTX)
PCR primer sequences.
(XLSX)
Acknowledgments
We are thankful to Nancy Reyes, Su Kah Goh and Deborah Davey for assistance with sample processing and experimentation.
Funding Statement
This project was supported through the 2010 round of the priority-driven Collaborative Cancer Research Scheme (grant 1002743) and funded by the Australian Government Department of Health and Ageing with the assistance of Cancer Australia. Also, this work was supported by the Australian National Health and Medical Research Council (grant numbers 145780, 288713; http://www.nhmrc.gov.au/); the Australian National Breast Cancer Foundation (grant PG-08-06; http://www.nbcf.org.au/); and the National Institutes of Health (grant 5U19AI067773-07). Support was also provided by the Victorian Government’s Operational Infrastructure Support Program. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Pierce AJ, Stark JM, Araujo FD, Moynahan ME, Berwick M, et al. (2001) Double-strand breaks and tumorigenesis. Trends in Cell Biology 11: S52–S59. [DOI] [PubMed] [Google Scholar]
- 2. Bernstein C, Bernstein H, Payne CM, Garewal H (2002) DNA repair/pro-apoptotic dual-role proteins in five major DNA repair pathways: fail-safe protection against carcinogenesis. Mutat Res 511: 145–178. [DOI] [PubMed] [Google Scholar]
- 3. Hoeijmakers JH (2009) DNA damage, aging, and cancer. N Engl J Med 361: 1475–1485. [DOI] [PubMed] [Google Scholar]
- 4. Tsuzuki T, Fujii Y, Sakumi K, Tominaga Y, Nakao K, et al. (1996) Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc Natl Acad Sci U S A 93: 6236–6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barnes DE, Stamp G, Rosewell I, Denzel A, Lindahl T (1998) Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr Biol 8: 1395–1398. [DOI] [PubMed] [Google Scholar]
- 6. Riballo E, Critchlow SE, Teo SH, Doherty AJ, Priestley A, et al. (1999) Identification of a defect in DNA ligase IV in a radiosensitive leukaemia patient. Curr Biol 9: 699–702. [DOI] [PubMed] [Google Scholar]
- 7. Abbaszadeh F, Clingen PH, Arlett CF, Plowman PN, Bourton EC, et al. (2010) A novel splice variant of the DNA-PKcs gene is associated with clinical and cellular radiosensitivity in a patient with xeroderma pigmentosum. J Med Genet 47: 176–181. [DOI] [PubMed] [Google Scholar]
- 8. Fishel R, Lescoe MK, Rao MR, Copeland NG, Jenkins NA, et al. (1993) The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell 75: 1027–1038. [DOI] [PubMed] [Google Scholar]
- 9. Papadopoulos N, Nicolaides NC, Wei YF, Ruben SM, Carter KC, et al. (1994) Mutation of a mutL homolog in hereditary colon cancer. Science 263: 1625–1629. [DOI] [PubMed] [Google Scholar]
- 10. Cleaver JE, Lam ET, Revet I (2009) Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nat Rev Genet 10: 756–768. [DOI] [PubMed] [Google Scholar]
- 11. Berneburg M, Lehmann AR (2001) Xeroderma pigmentosum and related disorders: defects in DNA repair and transcription. Adv Genet 43: 71–102. [DOI] [PubMed] [Google Scholar]
- 12. Lehmann AR (2003) DNA repair-deficient diseases, xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. Biochimie 85: 1101–1111. [DOI] [PubMed] [Google Scholar]
- 13. D'Andrea AD (2010) Susceptibility pathways in Fanconi's anemia and breast cancer. N Engl J Med 362: 1909–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wood RD, Mitchell M, Sgouros J, Lindahl T (2001) Human DNA repair genes. Science 291: 1284–1289. [DOI] [PubMed] [Google Scholar]
- 15. Wood RD, Mitchell M, Lindahl T (2005) Human DNA repair genes, 2005. Mutat Res 577: 275–283. [DOI] [PubMed] [Google Scholar]
- 16. Riches LC, Lynch AM, Gooderham NJ (2008) Early events in the mammalian response to DNA double-strand breaks. Mutagenesis 23: 331–339. [DOI] [PubMed] [Google Scholar]
- 17. Kis E, Szatmari T, Keszei M, Farkas R, Esik O, et al. (2006) Microarray analysis of radiation response genes in primary human fibroblasts. Int J Radiat Oncol Biol Phys 66: 1506–1514. [DOI] [PubMed] [Google Scholar]
- 18. Long XH, Zhao ZQ, He XP, Wang HP, Xu QZ, et al. (2007) Dose-dependent expression changes of early response genes to ionizing radiation in human lymphoblastoid cells. Int J Mol Med 19: 607–615. [PubMed] [Google Scholar]
- 19. Sprung CN, Li J, Hovan D, McKay MJ, Forrester HB (2011) Alternative transcript initiation and splicing as a response to DNA damage. PLoS ONE 6: e25758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ding LH, Shingyoji M, Chen F, Chatterjee A, Kasai KE, et al. (2005) Gene expression changes in normal human skin fibroblasts induced by HZE-particle radiation. Radiat Res 164: 523–526. [DOI] [PubMed] [Google Scholar]
- 21. Tachiiri S, Katagiri T, Tsunoda T, Oya N, Hiraoka M, et al. (2006) Analysis of gene-expression profiles after gamma irradiation of normal human fibroblasts. Int J Radiat Oncol Biol Phys 64: 272–279. [DOI] [PubMed] [Google Scholar]
- 22. Tsai MH, Chen X, Chandramouli GV, Chen Y, Yan H, et al. (2006) Transcriptional responses to ionizing radiation reveal that p53R2 protects against radiation-induced mutagenesis in human lymphoblastoid cells. Oncogene 25: 622–632. [DOI] [PubMed] [Google Scholar]
- 23. Severin DM, Leong T, Cassidy B, Elsaleh H, Peters L, et al. (2001) Novel DNA sequence variants in the hHR21 DNA repair gene in radiosensitive cancer patients. Int J Radiat Oncol Biol Phys 50: 1323–1331. [DOI] [PubMed] [Google Scholar]
- 24. Leong T, Whitty J, Keilar M, Mifsud S, Ramsay J, et al. (2000) Mutation analysis of BRCA1 and BRCA2 cancer predisposition genes in radiation hypersensitive cancer patients. Int J Radiat Oncol Biol Phys 48: 959–965. [DOI] [PubMed] [Google Scholar]
- 25. Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98: 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Paul S, Amundson SA (2008) Development of gene expression signatures for practical radiation biodosimetry. Int J Radiat Oncol Biol Phys 71: 1236–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rieger KE, Chu G (2004) Portrait of transcriptional responses to ultraviolet and ionizing radiation in human cells. Nucleic Acids Res 32: 4786–4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fousteri M, Mullenders LH (2008) Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res 18: 73–84. [DOI] [PubMed] [Google Scholar]
- 29. Amundson SA, Do KT, Shahab S, Bittner M, Meltzer P, et al. (2000) Identification of potential mRNA biomarkers in peripheral blood lymphocytes for human exposure to ionizing radiation. Radiat Res 154: 342–346. [DOI] [PubMed] [Google Scholar]
- 30. Jen KY, Cheung VG (2003) Transcriptional response of lymphoblastoid cells to ionizing radiation. Genome Res 13: 2092–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Riley PA (1994) Free radicals in biology: oxidative stress and the effects of ionizing radiation. Int J Radiat Biol 65: 27–33. [DOI] [PubMed] [Google Scholar]
- 32. Bohr VA, Dianov GL (1999) Oxidative DNA damage processing in nuclear and mitochondrial DNA. Biochimie 81: 155–160. [DOI] [PubMed] [Google Scholar]
- 33. Tanaka H, Arakawa H, Yamaguchi T, Shiraishi K, Fukuda S, et al. (2000) A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature 404: 42–49. [DOI] [PubMed] [Google Scholar]
- 34. Xue L, Zhou B, Liu X, Heung Y, Chau J, et al. (2007) Ribonucleotide reductase small subunit p53R2 facilitates p21 induction of G1 arrest under UV irradiation. Cancer Res 67: 16–21. [DOI] [PubMed] [Google Scholar]
- 35. Kimura T, Gotoh M, Nakamura Y, Arakawa H (2003) hCDC4b, a regulator of cyclin E, as a direct transcriptional target of p53. Cancer Sci 94: 431–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Batty D, Rapic'-Otrin V, Levine AS, Wood RD (2000) Stable binding of human XPC complex to irradiated DNA confers strong discrimination for damaged sites. J Mol Biol 300: 275–290. [DOI] [PubMed] [Google Scholar]
- 37. Buterin T, Meyer C, Giese B, Naegeli H (2005) DNA quality control by conformational readout on the undamaged strand of the double helix. Chem Biol 12: 913–922. [DOI] [PubMed] [Google Scholar]
- 38. Lee TI, Young RA (2000) Transcription of eukaryotic protein-coding genes. Annu Rev Genet 34: 77–137. [DOI] [PubMed] [Google Scholar]
- 39. Wang QE, Han C, Zhang B, Sabapathy K, Wani AA (2012) Nucleotide excision repair factor XPC enhances DNA damage-induced apoptosis by downregulating the antiapoptotic short isoform of caspase-2. Cancer Res 72: 666–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bunick CG, Miller MR, Fuller BE, Fanning E, Chazin WJ (2006) Biochemical and structural domain analysis of xeroderma pigmentosum complementation group C protein. Biochemistry 45: 14965–14979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Legerski R, Peterson C (1992) Expression cloning of a human DNA repair gene involved in xeroderma pigmentosum group C. Nature. 359: 70–73. [DOI] [PubMed] [Google Scholar]
- 42. Gonda DK, Bachmair A, Wunning I, Tobias JW, Lane WS, et al. (1989) Universality and structure of the N-end rule. J Biol Chem 264: 16700–16712. [PubMed] [Google Scholar]
- 43. Zauberman A, Flusberg D, Haupt Y, Barak Y, Oren M (1995) A functional p53-responsive intronic promoter is contained within the human mdm2 gene. Nucleic Acids Res 23: 2584–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B (1992) Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 358: 80–83. [DOI] [PubMed] [Google Scholar]
- 45. Barak Y, Gottlieb E, Juven-Gershon T, Oren M (1994) Regulation of mdm2 expression by p53: alternative promoters produce transcripts with nonidentical translation potential. Genes Dev 8: 1739–1749. [DOI] [PubMed] [Google Scholar]
- 46. Thut CJ, Goodrich JA, Tjian R (1997) Repression of p53-mediated transcription by MDM2: a dual mechanism. Genes Dev 11: 1974–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Radhakrishnan SK, Gierut J, Gartel AL (2006) Multiple alternate p21 transcripts are regulated by p53 in human cells. Oncogene 25: 1812–1815. [DOI] [PubMed] [Google Scholar]
- 48. Liu G, Chen X (2006) DNA polymerase eta, the product of the xeroderma pigmentosum variant gene and a target of p53, modulates the DNA damage checkpoint and p53 activation. Mol Cell Biol 26: 1398–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Amundson SA, Patterson A, Do KT, Fornace AJ Jr (2002) A nucleotide excision repair master-switch: p53 regulated coordinate induction of global genomic repair genes. Cancer Biol Ther 1: 145–149. [DOI] [PubMed] [Google Scholar]
- 50. Nakano K, Balint E, Ashcroft M, Vousden KH (2000) A ribonucleotide reductase gene is a transcriptional target of p53 and p73. Oncogene 19: 4283–4289. [DOI] [PubMed] [Google Scholar]
- 51. Xu J, Morris GF (1999) p53-mediated regulation of proliferating cell nuclear antigen expression in cells exposed to ionizing radiation. Mol Cell Biol 19: 12–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Modulation of DNA repair genes at the exon level 4 hours after treatment with 10 Gy IR in lymphoblast cell lines. PSR expression levels are plotted for each of the DNA repair genes with a p-value of <0.05 using Partek Genomics Suite statistical package. Relative fluorescence (y-axis; log base 2) is plotted for each PSR (x-axis). Core PSRs are labelled below the graphs. Samples were either sham irradiated (red) or irradiated (blue) with 10 Gy from a 137Cs source. Error bars = SEM (n = 12).
(PDF)
Modulation of DNA repair genes at the exon level 4 hours after treatment with 10 Gy IR in fibroblast cells. PSR expression levels are plotted for each of the DNA repair genes with a p-value of <0.05 using Partek Genomics Suite statistical package. Relative fluorescence (y-axis; log base 2) is plotted for each PSR (x-axis). Core PSRs are labelled below the graphs. Samples were either sham irradiated (red) or irradiated (blue) with 10 Gy from a 137Cs source. Error bars = SEM (n = 12).
(PDF)
Real-time PCR comparison of DNA repair gene expression between LCLs and fibroblasts. Initial levels of transcripts are significantly higher in fibroblasts (FB) for XPC and RRM2B relative to LCLs, but after IR induction, gene expression levels are not significantly different. Values have been normalized to PGK gene expression. The p-values for differences of initial levels between LCL and fibroblast samples are as follows: XPC: p = 0.0025 and for RRM2B: p = 0.0030.
(PPTX)
Sequence analysis of the 5′RACE amplicons for RRM2B (A) and XPC (B). Sequences homologous to the primers used for their amplifcation are indicated by long horizontal arrows below the sequence. The adaptor sequences are shown in capital letters. The vertical arrows indicate the start site for the capped mRNA transcript. This is nucleotide 35 of NCBI RRM2B sequence NM_001172477 (A), and at nucleotide 261 of NCBI XPC sequence NM_004628 (B).
(PPTX)
PCR primer sequences.
(XLSX)