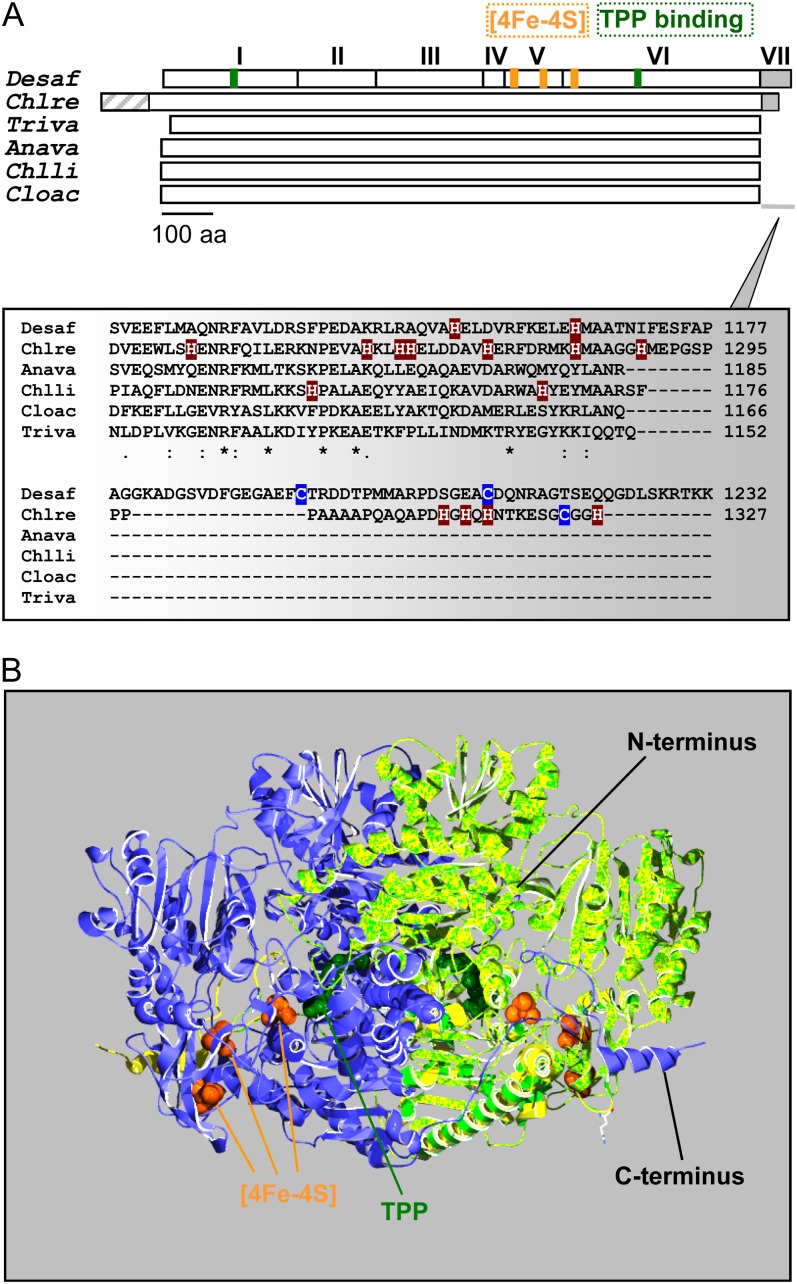
Figure 1.
Schematic representation and predicted structure of C. reinhardtii PFO. A, Top, the monomer of the homodimeric DaPFO has been described as being composed of seven domains (I–VII). Domains I, II, and IV are involved in dimer formation, and domain VII confers oxygen stability to the enzyme via disulfide bond formation (Pieulle et al., 1995; Chabrière et al., 1999). Bottom, as compared with bacterial and eukaryotic PFOs, the algal enzyme exhibits an extension at its N terminus that likely serves as an intracellular targeting peptide. The C-terminal extension of CrPFO differs from that of DaPFO in both length and amino acid (aa) composition, in particular in their contents of His and Cys residues. Anava, Anabaena variabilis (cyanobacterium); Chlli, Chlorobium limicola (chlorobia); Chlre, C. reinhardtii (photosynthetic eukaryote); Cloac, C. acetobutylicum (firmicute); Desaf, D. africanus (deltaproteobacterium); Triva, T. vaginalis (heterotrophic eukaryote). B, Overlay of the predicted structure of a CrPFO monomer (light green; Pro-96–His-1292) with the use of the DaPFO monomer (yellow) as a template shown in interaction with another DaPFO monomer (blue) to form the typical PFO homodimer. The 2.3-Å crystal structure of DaPFO has revealed the arrangement of the three [4Fe-4S] clusters (orange) and the TPP cofactor (dark green) within the enzyme (Chabrière et al., 1999; Charon et al., 1999). TPP is deeply buried in the protein, and its closest cluster (or proximal cluster) is approximately 13 Å away. The other two clusters are arranged successively up to the surface of the dimer, with cluster-to-cluster distances of about 12 to 15 Å. In DaPFO, the C terminus (domain VII) extends over the other monomer (Chabrière et al., 1999).