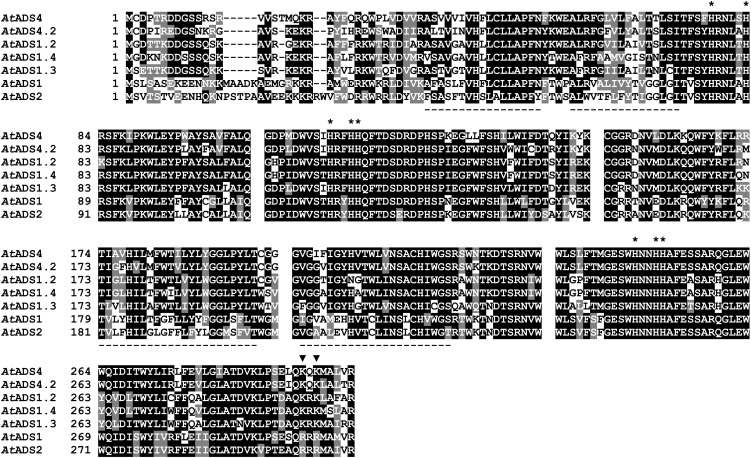
Figure 1.
Comparison of the deduced amino acid sequences of seven Arabidopsis ADS family members. Universal Protein Resource code numbers are given in Table I. Sequences were clustered using clustalW (http://www.genome.jp/tools/clustalw/) and colored using BOXSHADE 3.21 (http://www.ch.embnet.org/software/BOX_form.html). Gaps were inserted manually to reflect polypeptide sequences encoded by individual exons. His residues conserved in membrane-bound desaturases are marked by asterisks. Black triangles mark positions of residues potentially involved in ER retention/retrieval. Underlined sequences are predicted hydrophobic membrane spanning regions (http://www.cbs.dtu.dk/services/TMHMM-2.0/; www.predictprotein.org).