Figure 2.
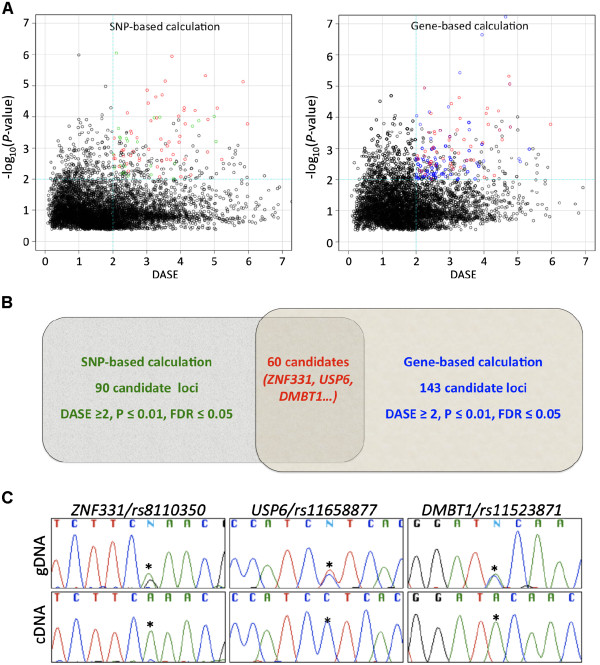
Identification of DASE loci. (A) We developed 2 statistical methods, SNP-based and gene-based calculation, to assign DASE values to transcribed loci. (B) Using the same selection stringency, sixty candidate loci were identified by both methods. Among them, ZNF331 and USP6 are classified as cancer causative by the Cancer Gene Census at Sanger Institute. The DMBT1 locus has been reported to contain breast cancer risk associated variants. (C) We used these three genes as examples to show our DASE validation process. Comparing sequencing chromagraphs between cDNA and gDNA amplicons we could easily tell a typical DASE pattern from all of them.
