Fig 4.
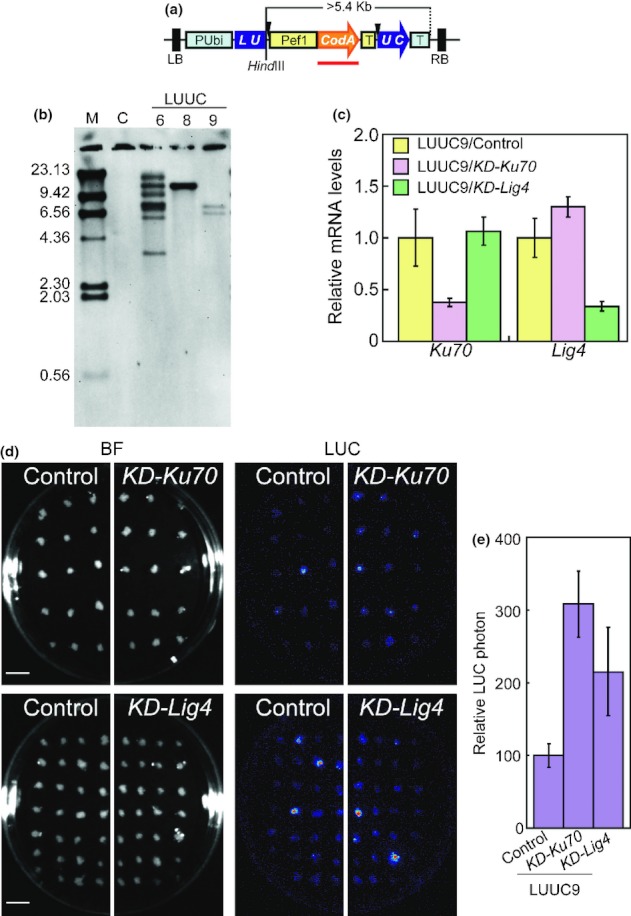
Luciferase (LUC) luminescence of recombination events by spontaneous double-strand breaks (DSBs) in control, KD-OsKu70 and KD-OsLig4 rice (Oryza sativa) calli. (a) Schematic diagram of the LU-UC recombination substrate. This recombination substrate consists of two partially duplicated fragments of the LUC gene interrupted by a cytosine deaminase (codA) expression cassette and two recognition sites for the meganuclease I-SceI (arrowheads). PUbi, maize ubiquitin1 promoter; Pef1, rice elongation factor 1 promoter; T, transcription terminator; LB, left border; RB, right border. Red bar represents DNA probe for Southern blot analysis. (b) Southern blot analysis using a codA probe (shown by the red bar in a) and 1 μg of genomic DNA extracted from wild-type and LU-UC recombination substrate transgenic rice calli and digested with HindIII. (c) Quantitative reverse transcriptase-polymerase chain reaction (RT-PCR) analysis of OsKu70 (left) and OsLig4 (right) transcripts in LU-UC-9 transgenic lines transformed with pANDA empty vector (as a control), KD-Ku70 or KD-Lig4. Relative transcript levels were normalized to OsAct1 mRNA. Error bars represent ± SD of three individual experiments. (d) Bright-field image (BF) and LUC luminescence image (LUC) of control, KD-Ku70 (upper panel) and KD-Lig4 (lower panel) rice calli. LUC luminescence derived from recombination events of recombination substrate by spontaneous DSB were detected in 4-wk-old transgenic rice calli without induction of I-SceI expression. Bar, 1 cm. (e) Graphical representation of the LUC luminescence intensity on control and KD-Ku70 and KD-Lig4 rice calli. Relative LUC luminescence levels were normalized to LUC luminescence levels of control calli (= 100). Error bars represent ± SD.
