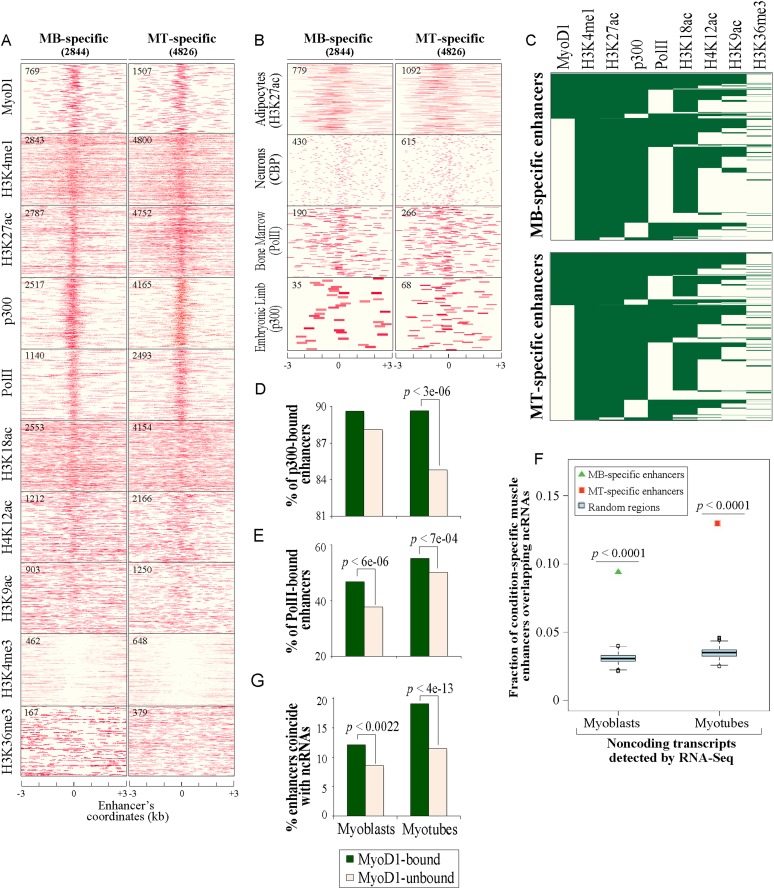
Figure 3.
Muscle enhancers are uniquely associated with spatially constrained chromatin marks. (A) Chromatin state maps of enhancer-related markers and several other histone marks within a region ±3 kb of the center of condition-specific enhancers. Data for MyoD1-binding events were obtained from Cao et al. (2010). p300- and H3K27ac-binding events (enriched tags) were identified using Qeseq (Supplemental Material). ChIP-seq data for p300 and H3K27ac were generated in this study, and all other data were published previously (Asp et al. 2011). The number of condition-specific enhancers significantly enriched with the indicated mark is indicated in the top left corner of each map. (B) Correspondence between muscle enhancers and enhancers in nonmuscle tissues. Enriched binding events of enhancer-related marks obtained from several nonmuscle mouse tissues (Visel et al. 2009; De Santa et al. 2010; Kim et al. 2010; Mikkelsen et al. 2010) corresponding to condition-specific muscle enhancers are plotted as indicated. (C) Clustering of MyoD1, enhancer features, and other histone marks based on their deposition at condition-specific muscle enhancers. (D,E) Recruitment of p300 and Pol II to MyoD1-bound enhancers. Recruitment of p300 (D) and Pol II (E) is significantly higher on enhancers bound by MyoD1 (two-proportion z-test). (F) Enrichment of condition-specific enhancers with transcribed ncRNAs deduced from RNA-seq (Trapnell et al. 2010). ncRNAs were overlapped with condition-specific enhancers or random data sets (Supplemental Material). The fractions of condition-specific enhancer and the respective distributions of random, control sequences (cyan box plots) overlapping ncRNAs are presented. (G) Coincidence of ncRNAs with enhancers is significantly higher on MyoD1-bound enhancers.