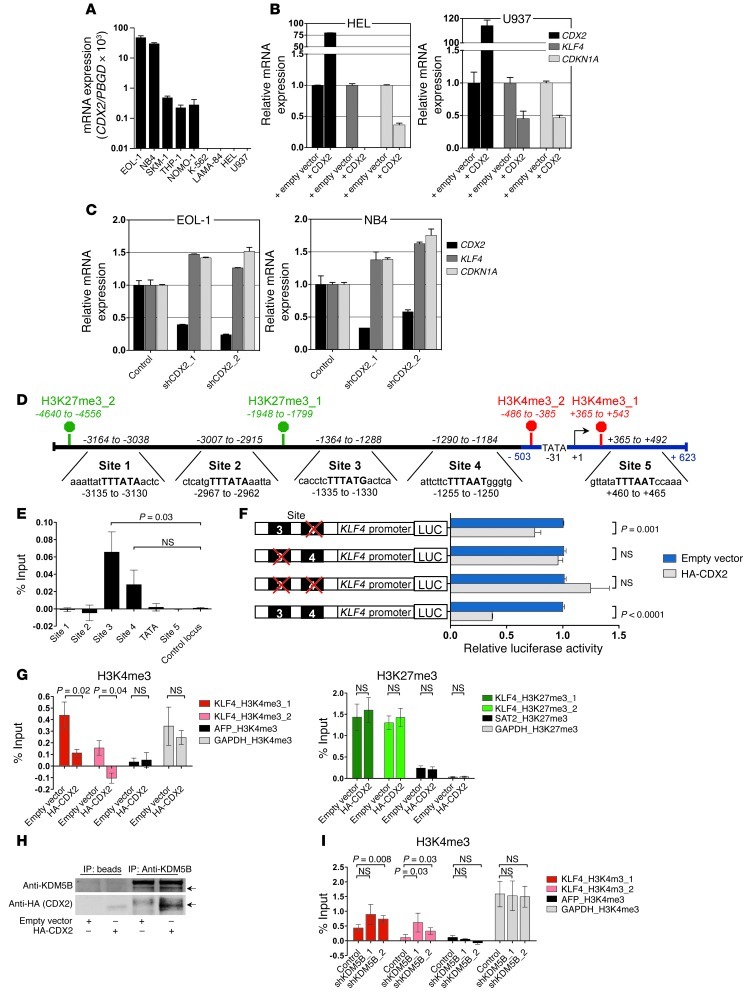
Figure 2. Relationship between CDX2 and KLF4 expression and analysis of the KLF4 regulatory region in human myeloid leukemia cell lines.
(A) CDX2 mRNA expression. (B) KLF4 and CDKN1A mRNA levels in response to ectopic CDX2 expression. (C) KLF4 and CDKN1A mRNA levels in response to CDX2 knockdown. (D) Predicted KLF4 promoter (blue) and 4.6 kb upstream of the KLF4 TSS (+1). Possible CDX2 binding sites and the location of the corresponding PCR amplicons (italics) used for ChIP are indicated. The consensus sequence for binding of Cdx proteins is given in bold capital letters. H3K4me3 and H3K27me3 marks, detected in K-562 cells in the ENCODE Histone Modification ChIP-seq project by the Broad Institute, are indicated in red and green, respectively. (E) ChIP showing occupancy of sites 3 and 4 by HA-CDX2 in K-562. GAPDH served as control locus. (F) LUC reporter assays showing that site 3 is required for transcriptional silencing of KLF4 in response to ectopic CDX2 expression in K-562. Mutated sites are indicated by red crosses. (G) ChIP showing reduced H3K4me3 enrichment and unchanged H3K27me3 occupancy in K-562 upon expression of HA-CDX2. AFP and GAPDH served as negative and positive control loci for H3K4me3; GAPDH and SAT2 served as negative and positive control loci for H3K27me3. (H) Co-IP demonstrating interaction of HA-CDX2 with KDM5B in K-562. (I) ChIP showing increased H3K4me3 enrichment in K-562 stably transduced with HA-CDX2 upon KDM5B knockdown. AFP and GAPDH served as negative and positive control loci.