Abstract
Late-stage breast cancer metastasis is driven by dysregulated TGF-β signaling, but the underlying molecular mechanisms have not been fully elucidated. We attempted to recapitulate tumor and metastatic microenvironments via the use of biomechanically compliant or rigid 3D organotypic cultures and combined them with global microRNA (miR) profiling analyses to identify miRs that were upregulated in metastatic breast cancer cells by TGF-β. Here we establish miR-181a as a TGF-β–regulated “metastamir” that enhanced the metastatic potential of breast cancers by promoting epithelial-mesenchymal transition, migratory, and invasive phenotypes. Mechanistically, inactivation of miR-181a elevated the expression of the proapoptotic molecule Bim, which sensitized metastatic cells to anoikis. Along these lines, miR-181a expression was essential in driving pulmonary micrometastatic outgrowth and enhancing the lethality of late-stage mammary tumors in mice. Finally, miR-181a expression was dramatically and selectively upregulated in metastatic breast tumors, particularly triple-negative breast cancers, and was highly predictive for decreased overall survival in human breast cancer patients. Collectively, our findings strongly implicate miR-181a as a predictive biomarker for breast cancer metastasis and patient survival, and consequently, as a potential therapeutic target in metastatic breast cancer.
Introduction
Metastasis is a complex multistage process whereby primary tumor cells acquire the ability to (a) locally invade through the surrounding stroma; (b) intravasate into blood vessels; (c) survive transit through the vascular system; (d) extravasate and arrest at distant sites; and (e) survive in foreign microenvironments and overcome systemic dormancy to undergo metastatic outgrowth, ultimately leading to the formation of secondary tumors in vital organ sites (1). Metastasis of primary mammary tumors accounts for the vast majority of deaths of breast cancer patients. Indeed, the 5-year survival rate for patients with breast cancer drops precipitously from 98% for individuals with localized disease to 23% for those with metastatic disease (2). Within normal mammary tissues, the multifunctional cytokine TGF-β functions as a potent tumor suppressor through its ability to induce cell-cycle arrest and apoptosis. Unlike their normal counterparts, malignant mammary tissues can transform the normal functions of TGF-β to that of a potent stimulator of breast cancer proliferation, migration, and invasion in part via its ability to promote the acquisition of epithelial-mesenchymal transition (EMT) and metastatic phenotypes (3–5). This switch in TGF-β function from that of a tumor suppressor to a tumor promoter is known as the “TGF-β paradox”; the mechanistic underpinnings that engender this phenomenon remain incompletely understood. Moreover, this switch in TGF-β function is often accompanied by desmoplastic and fibrotic reactions, which elicit dramatic changes in the biomechanical properties of the tumor microenvironment. Indeed, the elastic modulus of stroma housed within breast carcinomas is approximately 10 times more mechanically rigid than that of adjacent normal breast tissues (6, 7). TGF-β potentiates these biomechanical reactions by stimulating the expression and secretion of a variety of ECM components, such as collagen I and fibronectin from stromal fibroblasts, and of ECM cross-linking enzymes, such as lysyl oxidase from mammary carcinoma cells (3, 4, 8). The formation of these rigid mammary tumor microenvironments promotes metastatic progression in breast cancers and also predicts poor clinical outcomes in patients harboring metastatic disease (6, 9–12). Interestingly, normal mammary and lung tissues share similarly compliant elastic moduli, a biomechanical condition that may contribute to initiation of dormancy by disseminated breast micrometastases in the lungs (13). We recently demonstrated that biomechanically compliant microenvironments can reinstate the cytostatic activities of TGF-β in late-stage breast cancer cells, indicating that matrix rigidity plays a vital role in mediating how cells sense and respond to the dichotomous functions of TGF-β (8). Moreover, the ability of carcinoma cells to thrive both in rigid primary tumor microenvironments and compliant metastatic microenvironments represents an essential characteristic of fully metastatic breast cancer cells. It therefore stands to reason that enhancing our knowledge of the molecular mechanisms that mediate breast cancer metastasis may enable the development of specific metastasis-based treatments needed to improve the overall survival rates of patients harboring metastatic breast cancers.
MicroRNAs (miRs) are small (20–30 nucleotides) noncoding RNAs that posttranscriptionally regulate gene expression through canonical base pairing between the miR seed sequence (nucleotides 2–8 of the 5′ end) and the complementary sequence in the 3′ UTR of the target mRNA. The net effect of these events elicits either translational repression or degradation of targeted mRNAs (14). Recently, several studies have implicated aberrant miR expression in the development and metastatic progression of mammary tumors (15). At present, the precise role of miRs in controlling metastatic progression by TGF-β remains to be fully elucidated, as does the impact of tissue compliance in altering these TGF-β– and miR-driven activities. Global miR expression profiling analyses enabled us to identify a variety of miRs whose expression is regulated by TGF-β and altered ECM rigidity, one of which was miR-181a, which is upregulated by TGF-β in hepatocytes (16, 17) and breast cancer cells (15). Importantly, we demonstrate that aberrantly high miR-181a expression enhanced the ability of TGF-β to stimulate breast cancer metastasis by inducing EMT programs and by promoting resistance to anoikis by downregulating the expression of the proapoptotic factor Bim. Equally important, miR-181a expression is highly associated with the development of metastatic disease in breast cancers, particularly triple-negative breast cancers (TNBCs), and is highly predictive for poor clinical outcomes in breast cancer patients. Taken together, our findings establish miR-181a as a potential predictive biomarker for breast cancer metastasis and overall survival as well as a promising pharmacologic target for the treatment of metastatic breast cancers.
Results
miR-181a expression is upregulated by TGF-β and correlates with the metastatic potential of breast cancer cells.
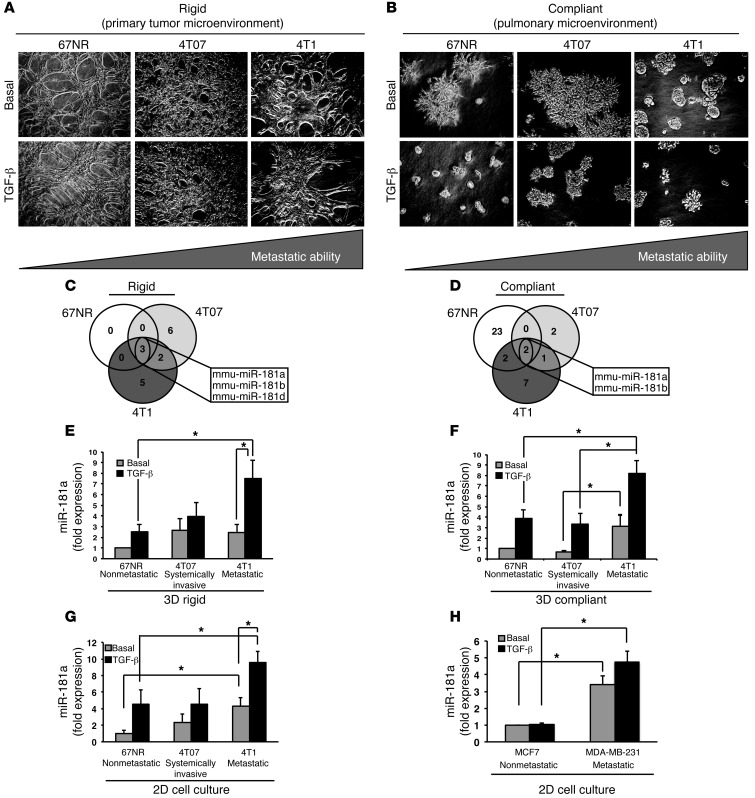
Several studies have recently depicted the role of TGF-β in upregulating the expression of several oncogenic miRs via transcriptional and miR processing mechanisms (18–20). Along these lines, we recently demonstrated the ability of biomechanically rigid ECM to regulate the differential responses of normal versus malignant mammary epithelial cells (MECs) to TGF-β (8). Likewise, increasing the rigidity of tumor microenvironments not only drives breast cancer invasion and metastasis, but also diminishes the effective delivery and penetration of chemotherapeutic agents, thereby safeguarding the survival of the developing tumor (13, 21). However, the role of ECM rigidity in dictating miR expression governed by TGF-β remains undefined. As a means to fill this knowledge gap, we utilized 3D organotypic cultures whose physical properties were altered by inclusion of type I collagen to create biomechanically rigid microenvironments that approximated those typically observed in primary mammary tumors (Figure 1A and refs. 8, 13, 21). Compliant 3D organotypic cultures (i.e., no collagen supplementation) were also generated to recapitulate the biomechanical properties of pulmonary microenvironments typically encountered by disseminated breast carcinoma cells (Figure 1B and refs. 8, 22). The murine 4T1 progression series represents an established model of TNBC development and metastasis and consists of isogenically derived nonmetastatic 67NR, systemically invasive 4T07, and highly metastatic 4T1 cells (23) that were propagated for 6 days in the absence or presence of TGF-β in either rigid or compliant 3D cultures (Figure 1, A and B). Afterward, total RNA was extracted and subjected to miR profiling. It is important to note that the ensuing discussion only relates to miRs whose expression was significantly induced by TGF-β. The presentation and discussion of miRs whose expression was significantly downregulated by TGF-β will be presented elsewhere. As such, Figure 1, C and D, clearly show that the coupling of TGF-β to the induction of miR expression was highly dependent upon tissue compliance. For instance, when propagated under biomechanically rigid 3D organotypic cultures, TGF-β significantly induced the expression of 3 miRs in 67NR cells, 11 miRs in 4T07 cells, and 10 miRs in 4T1 (Supplemental Tables 1–3; supplemental material available online with this article; doi: 10.1172/JCI64946DS1). In stark contrast, administering TGF-β to these same cells when propagated in biomechanically compliant 3D organotypic cultures significantly stimulated the expression of 27, 5, and 12 miRs in 67NR, 4T07, and 4T1 cells, respectively (Figure 1D and Supplemental Tables 1–3). Interestingly, only miRs belonging to the miR-181 family were universally upregulated by TGF-β in all 3 isogeneic cell lines and 3D organotypic culture conditions, suggesting that TGF-β functions as a master regulator of this miR family (Figure 1, C and D). Accordingly, examining the expression levels of miR-181a by semiquantitative real-time PCR demonstrated that TGF-β was significantly more effective in stimulating miR-181a expression in metastatic 4T1 cells as compared with their nonmetastatic 67NR counterparts. Importantly, the coupling of TGF-β to miR-181a expression readily transpired in both rigid and compliant 3D organotypic cultures (Figure 1, E and F) as well as in traditional 2D culture systems (Figure 1G). Moreover, these analyses showed that the magnitude of miR-181a expression stimulated by TGF-β correlated positively with the metastatic potential of these isogeneic derivatives (Figure 1, E–G). To ensure that these findings were not limited to the 4T1 progression series, we also compared the ability of TGF-β to induce the expression of miR-181a in human MCF-7 (i.e., luminal and nonmetastatic) and MBA-MD-231 (i.e., triple-negative and metastatic) breast cancer cells. Although MCF-7 cells respond to TGF-β (Supplemental Figure 1 and ref. 24), miR-181a expression remained low and unresponsive to TGF-β in MCF-7 cells, an event that was in stark contrast to the significant induction of miR-181a expression in MDA-MB-231 cells both prior and after their stimulation with TGF-β (Figure 1H). Recently, MCF-7 cells were observed to downregulate miR-181a in response to estrogen administration (25), and this finding together with those presented herein suggests that the induction of miR-181a expression by TGF-β may be selectively sustained in metastatic TNBCs. Accordingly, TGF-β treatment of nonmetastatic cells over a span of 48 hours led to a transient increase in miR-181a expression that peaked at 16 hours and thereafter declined during the ensuing time points to 48 hours (Supplemental Figure 2, A and B). In stark contrast, miR-181a expression was progressively increased and sustained in metastatic TNBCs upon completion of TGF-β treatment (Supplemental Figure 2, A and B). Thus, metastatic human and murine breast cancers stably upregulate miR-181a in response to TGF-β. Interestingly, conflicting reports in the literature show that TGF-β–mediated upregulation of miR-181a occurs through either transcriptional (16) or posttranslational mechanisms (15). Moreover, miR-181a is transcribed from 2 separate genomic loci that give rise to 2 separate pre-miRs, namely pre-miR-181a-1 and pre-miR-181a-2 ( http://www.mirbase.org/). Our own analyses indicate that the transcription of pre-miR-181a-1 transpires through a Smad4-dependent mechanism, while that of pre-miR-181a-2 occurs independently of Smad4 (Supplemental Figure 3, A and B). However, levels of mature miR-181a are not affected by Smad4 depletion (Supplemental Figure 3C), indicating that Smad4-independent processing steps are likely to occur after production of the pre-miR-181a transcripts.
Figure 1. miR-181a expression is upregulated by TGF-β and correlates with the metastatic potential of breast cancer cells.
(A and B) Nonmetastatic 67NR, systemically invasive 4T07, and broadly metastatic 4T1 cells were propagated in the absence or presence of TGF-β1 (5 ng/ml) for 6 days in either rigid (3 mg/ml collagen) or compliant 3D organotypic cultures. Total RNA was harvested and hybridized to miRIDIAN miR arrays by Dharmacon. Original magnification, ×20. (C and D) Venn diagrams depicting miRs induced by TGF-β in 67NR, 4T07, and 4T1 cells revealed that the miR-181 family was upregulated by TGF-β1 treatment in all breast cancer cell lines and treatment conditions. mmu, Mus musculus. (E and F) TGF-β1 (5 ng/ml) treatment of 67NR, 4T07, and 4T1 cells propagated in either rigid (3 mg/ml collagen, E) or compliant (F) 3D organotypic cultures increased miR-181a expression in a manner correlated with the metastatic potential of individual breast cancer cell lines. (G and H) TGF-β1 (5 ng/ml) treatment of mouse 67NR, 4T07, and 4T1 (G), and of human MCF-7 and MDA-MB-231 (H) cells propagated in traditional 2D cultures stimulated miR-181a expression in a manner correlated with the metastatic potential of the individual cell lines. Individual miR signals were normalized to U6, and the data are presented as the mean ± SEM fold expression of miR-181a relative to basal 67NR or MCF-7 cells. n = 3. *P < 0.05, Student’s t test.
TGF-β has previously been reported to regulate the expression of miRs 181a and 181b in hepatocytes (16, 17) and perhaps in cancer initiating cells of the breast as well (15). Accordingly, TGF-β significantly upregulated miR-181b expression in the 4T1 progression series in both rigid and compliant 3D organotypic cultures, in traditional 2D cultures, and predominantly in metastatic MDA-MB-231 cells as compared with nonmetastatic MCF-7 cells (Supplemental Figure 4, A and B, and Supplemental Figure 5, A and B). Moreover, the extent to which TGF-β coupled to the expression of the remaining miR-181 family members, namely miRs 181c and 181d, generally trended with those delineated for their miR 181a and 181b counterparts (Supplemental Figure 4, C–F, and Supplemental Figure 5, C–F). Collectively, these findings identify TGF-β as a master regulator of the expression of miR-181 family members and also implicate the expression of these miRs as potential mediators and biomarkers of breast cancer metastasis, particularly that in TNBCs.
Inactivation of miR-181a attenuates TGF-β–mediated EMT, invasion, and migration.
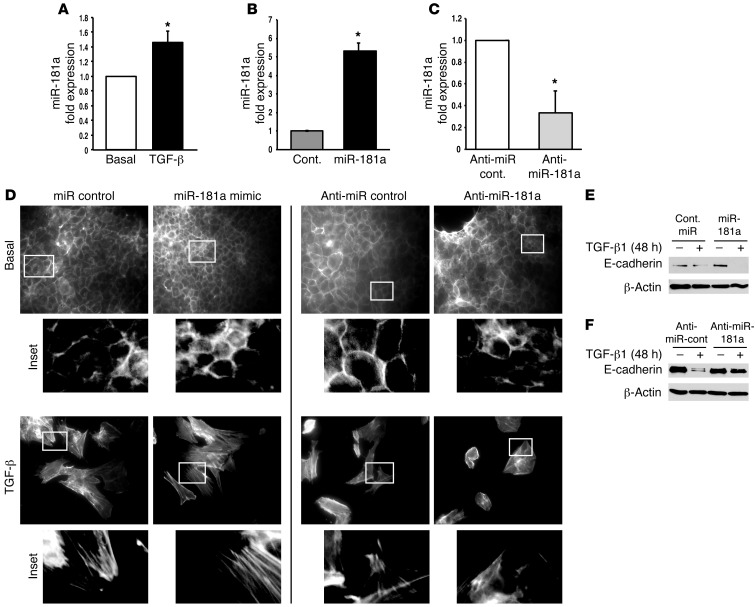
Given the parallels between miR-181a expression and metastatic potential, we next sought to investigate the probable role of miR-181a during EMT programs, which are critically involved in several aspects of metastasis stimulated by TGF-β (3). In doing so, we utilized normal murine mammary gland (NMuMG) cells, which are used routinely as a model system for studying the molecular mechanisms whereby TGF-β promotes EMT (8, 26–28). Figure 2A shows that TGF-β treatment of NMuMG cells under experimental conditions known to induce EMT programs resulted in the significant upregulation of miR-181a expression. Interestingly, although the expression of miR-181a Mimics in NMuMG cells (Figure 2B) failed to alter their ability to remodel the actin cytoskeletal system during EMT reactions (Figure 2D), we did observe the expression of miR-181a antagonists (Figure 2C) to attenuate the formation of actin stress fibers in NMuMG cells stimulated with TGF-β (Figure 2D). Along these lines, miR-181a Mimics enhanced the extent to which TGF-β downregulated E-cadherin expression in transitioning NMuMG cells (Figure 2E). Likewise, inactivation of miR-181a in NMuMG cells prevented the loss of E-cadherin expression stimulated by TGF-β (Figure 2F). Taken together, these results indicate that miR-181a participates in mediating the induction of EMT programs stimulated by TGF-β.
Figure 2. Inhibition of miR-181a attenuates TGF-β–mediated EMT in normal MECs.
(A) NMuMG cells were stimulated to undergo EMT with TGF-β1 (5 ng/ml), which upregulated miR-181a expression as determined by semiquantitative real-time PCR, where individual miR signals were normalized to U6. Data are the mean ± SEM fold expression relative to basal cells. n = 3. *P < 0.05, Student’s t test. (B and C) Transient transfection of miR-181a Mimics (B) or Hairpin inhibitors (C) resulted in elevated or diminished miR-181a expression in NMuMG cells. Data are the mean ± SEM fold expression relative to corresponding controls. n = 3. *P < 0.05, Student’s t test. (D–F) miR-181a expression levels were manipulated in NMuMG cells as above and subsequently stimulated with TGF-β1 (5 ng/ml) for 48 hours to induce an EMT program, which was monitored by phalloidin staining to visual alterations in the actin cytoskeleton (D) or by immunoblotting to monitor E-cadherin expression (E and F). Inactivation of miR-181a (Anti miR-181a) blunted TGF-β stimulation of EMT programs in NMuMG cells. Original magnification, ×20; insets, ×10 magnification of the original. All are representative findings observed in 3 independent experiments.
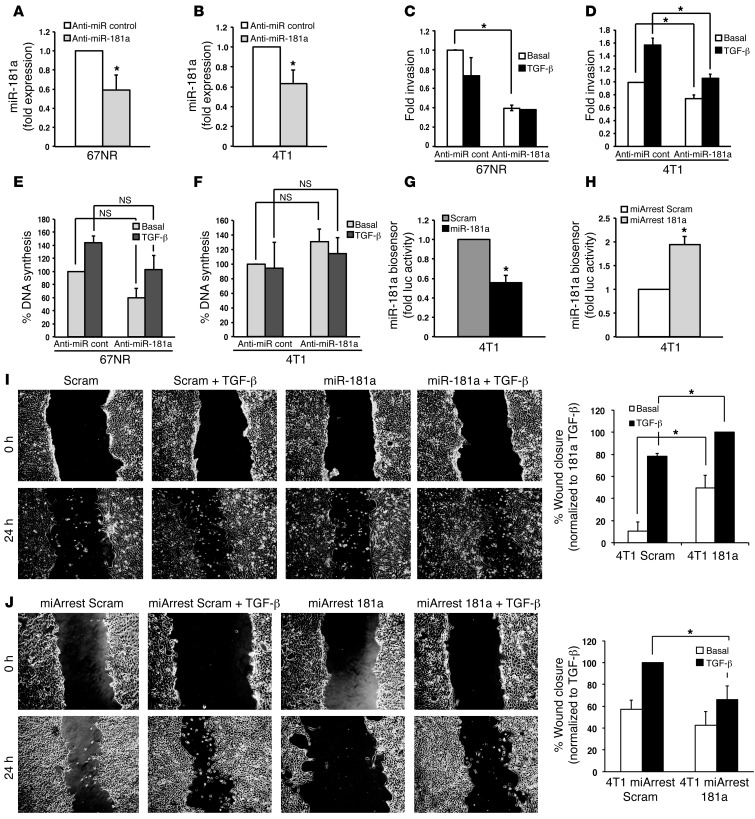
An essential manifestation of EMT programs is their ability to confer transitioned cells with highly motile and invasive phenotypes, and so we investigated the impact of manipulating miR-181a activity on the coupling of TGF-β to these cellular processes in nonmetastatic 67NR (Figure 3A) and metastatic 4T1 (Figure 3B) cells. Figure 3, C and D, shows that inactivating miR-181a decreased the ability of either cell type to invade to a serum stimulus. Indeed, nonmetastatic 67NR cells failed to invade when stimulated by TGF-β, a response that was unaffected by alterations in miR-181a activity (Figure 3C). However, inactivating miR-181a in metastatic 4T1 cells significantly impaired their ability to acquire invasive phenotypes in response to TGF-β (Figure 3D). Importantly, the reduced capacity of these cell lines to invade reconstituted basement membranes was not due to differences in their rates of DNA synthesis (Figure 3, E and F). Because expression of miR-181a Mimics failed to augment basal and TGF-β–stimulated invasion in either 67NR or 4T1 cells (Supplemental Figure 6, A and B), these findings suggest that the expression of miR-181 is necessary but not sufficient in driving breast cancer invasion stimulated by TGF-β.
Figure 3. Inhibition of miR-181a attenuates TGF-β–mediated EMT, invasion, and migration.
(A and B) An antagomir against miR-181a (anti–miR-181a) was transiently transfected into 67NR or 4T1 cells, resulting in decreased expression of miR-181a, as determined by semiquantitative real-time PCR. (C and D) The invasiveness of miR-181a–manipulated 67NR and 4T1 cells in response to TGF-β1 (5 ng/ml) treatment was significantly reduced by miR-181a inactivation. (E and F) The proliferation of miR-181a–manipulated 67NR and 4T1 cells in response to TGF-β1 (5 ng/ml) treatment was unaffected by miR-181a inactivation. (G and H) 4T1 cells engineered to stably express miR-181a (G) or a miR-181a sponge (H) were transiently transfected with a renilla luciferase miR-181a biosensor and CMV–β-gal, which was used to control for differences in transfection efficiency. miR-181a was shown to significantly elevate miR-181a activity, while miR-181a antagonists were shown to significantly reduce miR-181a activity. (I and J) The ability of TGF-β1 (5 ng/ml) to induce the migration of 4T1 cells with elevated (I) or reduced (J) miR-181a activity was significantly stimulated by miR-181a activation (I) or was significantly inhibited by miR-181a inactivation (J). Original magnification, ×20. All data are the mean ± SEM. n = 3. *P < 0.05, Student’s t test.
We also engineered 4T1 cells to stably express miR-181a, whose functionality was confirmed by detecting decreased renilla luciferase activity driven by a miR-181a biosensor (Figure 3G). Likewise, stable antagonism of miR-181a (i.e., miArrest 181a expression) significantly increased the activity of the miR-181a biosensor in 4T1 cells, a finding indicative of diminished miR-181a activity in these metastatic breast cancer cells (Figure 3H). Importantly, we observed elevated miR-181a activity as significantly increasing both basal and TGF-β–stimulated wound closure (Figure 3I), while the converse manipulation of miR-181a activity abrogated cell migration stimulated by TGF-β (Figure 3J). We also found stable increases or decreases in miR-181a activity to be incapable of affecting 4T1 proliferation in traditional 2D culture systems (data not shown). Collectively, these findings indicate that the upregulated expression of miR-181a enhances the motility and invasion of breast cancer cells, presumably by augmenting the coupling of TGF-β to the induction of EMT programs (Figure 3).
miR-181a expression enhances Erk1/2, Akt, and Src signaling in breast cancer cells.
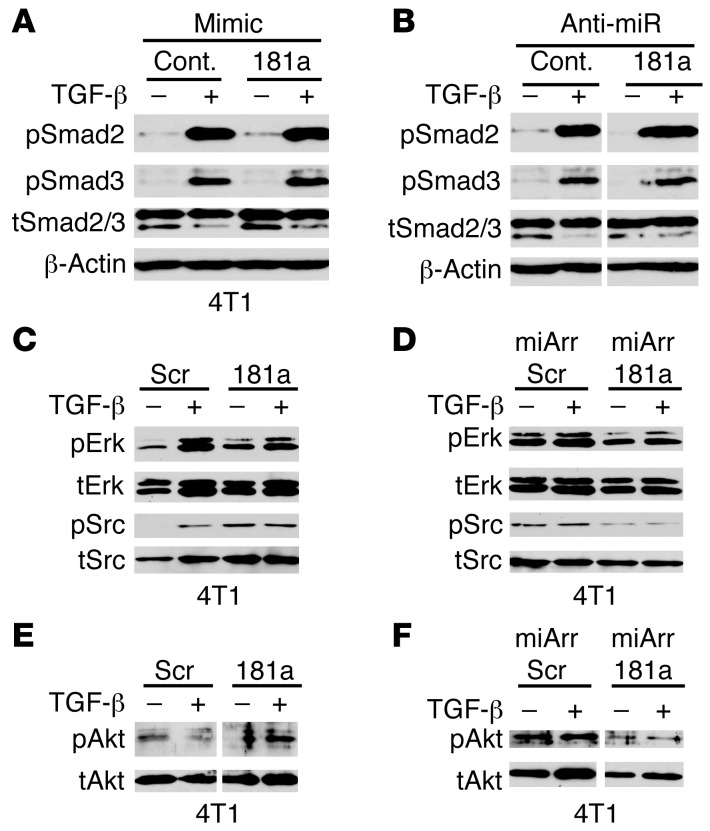
We next addressed whether changes in miR-181a activity were capable of altering the coupling of TGF-β to its downstream effectors, particularly those coupled to metastatic progression. Transmembrane signaling by TGF-β transpires following its activation of canonical Smad2/3/4 signaling, as well as its stimulation of a variety of noncanonical effectors, including MAP kinases, PI3K/Akt, and NF-κB (3–5, 29). The coupling of TGF-β to canonical Smad-based signaling is generally associated with the tumor-suppressing functions of TGF-β and predominates in normal MECs, which contrasts sharply with malignant MECs and their amplification of noncanonical TGF-β signaling that underlies its oncogenic functions in late-stage breast cancers (4). Consistent with this model, we failed to observe any changes in the ability of TGF-β to activate Smads 2 and 3 in miR-181a–manipulated 4T1 cells (Figure 4, A and B). Thus, alterations in miR-181a expression do not affect the initial activation of canonical signaling by TGF-β. Interestingly, elevating the activity of miR-181a enhanced the basal phosphorylation status of Src, Akt, and Erk1/2 (Figure 4, C and E). Likewise, diminishing the activity of miR-181a decreased both the basal and TGF-β–stimulated phosphorylation of Src, Akt, and Erk1/2 in 4T1 cells (Figure 4, D and F). Taken together, these findings suggest that elevated miR-181a functions to enhance the autoactivation of noncanonical TGF-β effectors, thereby mimicking oncogenic TGF-β signaling in metastatic breast cancer cells.
Figure 4. miR-181a expression enhances Erk1/2, Akt, and Src signaling in breast cancer cells.
(A–F) 4T1 cells that harbored manipulated miR-181a activity as indicated were stimulated with TGF-β1 (5 ng/ml) for 30 minutes. Afterward, the phosphorylation and expression of Smad2/3, Erk1/2, Akt, and Src were measured by immunoblotting as indicated. Shown are representative images from 3 independent experiments. Scr, scrambled control vector; miArr, miArrest vector. Lanes in B, E, and F were run on the same gel, but were noncontiguous (white lines).
miR-181a inhibits anoikis by targeting the proapoptotic protein Bim for downregulation.
The ability to overcome anoikis is paramount in promoting hematogenous dissemination of carcinoma cells (30). We therefore hypothesized upregulated miR-181a activity as a potential player in conferring breast cancer cell resistance to anoikis. We tested this supposition by culturing NMuMG and 4T1 cells over polyhydroxyethylmethacrylate (poly-HEMA) to prevent their adherence to plastic, thereby eliciting anoikis reactions. Figure 5A shows that parental NMuMG cells readily initiated apoptosis and caspase-3 cleavage in as little as 4 hours following adhesion deprivation. Importantly, elevating miR-181a activity completely inhibited the cleavage of caspase-3 in NMuMG cells that were suspended over a span of 24 hours (Figure 5A), indicating that miR-181a expression was sufficient to abrogate anoikis in NMuMG cells. Accordingly, inhibiting miR-181a expression in 4T1 cells sensitized these carcinoma cells to more rapidly undergo anoikis when deprived of adherence as compared with their parental counterparts (Figure 5B). The administration of TGF-β during this process failed to have a significant impact on the induction of anoikis (Supplemental Figure 7, A and B). However, pretreating 4T1 cells with the small molecule TβR-I antagonist TβR-I inhibitor II abrogated the high levels of autocrine TGF-β signaling in these cells (8, 26, 31, 32), thereby accelerating the rate of caspase-3 cleavage during anoikis in parental 4T1 cells (Supplemental Figure 7C). Importantly, miR-181a overexpression partially protected 4T1 cells from exhibiting accelerated caspase-3 cleavage and anoikis elicited by administration of TβR-II inhibitor II (Supplemental Figure 7D). Thus, TGF-β and miR-181a mediate the survival of nonadherent metastatic breast cancer cells.
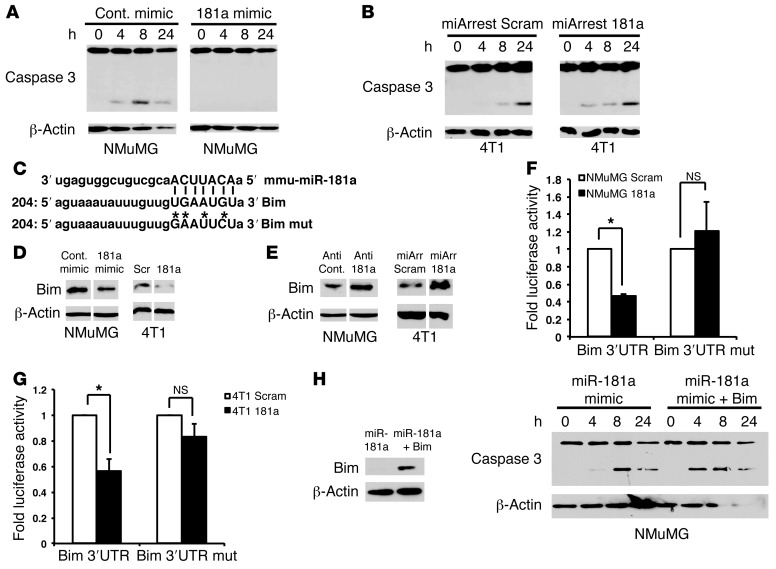
Figure 5. miR-181a inhibits anoikis by targeting the proapoptotic protein Bim for downregulation.
(A and B) NMuMG and 4T1 cells with manipulated miR-181a activity were suspended over poly-HEMA–coated culture dishes for 0–48 hours to induce anoikis. Caspase-3 cleavage was monitored by immunoblotting detergent-solubilized whole-cell extracts with anti–caspase-3 antibodies, while differences in protein loading were controlled with anti–β-actin antibodies. (C) miRanda alignment of mmu-miR-181a to the wild-type and mutant Bim 3′ UTR sequences demonstrates perfect complementation between the miR-181a seed sequence and the Bim 3′UTR. Asterisks indicate mutated miR-181a seed sequence binding bases. (D and E) Immunoblotting NMuMG and 4T1 cell extracts demonstrated that elevated miR-181a activity decreased Bim protein levels (D), while diminished miR-181a activity elicited increased Bim protein levels. Differences in protein loading were assessed by β-actin immunoblotting. (F and G) NMuMG (F) or 4T1 (G) cells were transiently transfected with a renilla luciferase reporter gene whose expression was driven by either wild-type or mutant (mut) Bim–3′ UTR seed sequences (C). Stable miR-181a expression suppressed that of luciferase containing the wild-type Bim–3′ UTR, an event that was lacking in cells transfected with mut–Bim–3′ UTR vectors. (H) NMuMG cells were transiently transfected with miR-181a Mimics with or without a 3′ UTR–deficient Bim cDNA. Afterward, the transfectants were suspended over poly-HEMA–coated culture dishes for 24 hours to induce anoikis. Bim expression (left panel) and caspase-3 cleavage (right panel) were monitored as described above. All data are representative of 3 independent experiments or are the mean ± SEM. n = 3; *P < 0.05; Student’s t test. Lanes in D and E were run on the same gel, but were noncontiguous (white lines).
To identify mRNA targets of miR-181a that are relevant to anoikis, we interrogated TargetScan (33) and miRanda (34) miR target prediction programs in combination with Ingenuity Pathway Analysis. In doing so, we identified the proapoptotic BH3-only protein Bim as a possible target of miR-181a. Bim promotes apoptosis by binding to the prosurvival proteins Bcl-2 and Bcl-XL, thereby engendering the release of Bax and Bak necessary to initiate programmed cell death (35). miR-181a has previously been reported to target several Bcl-2 family members in astrocytes, glioblastomas, and hematologic malignancies (36–38). Moreover, the 3′ UTR of Bim houses a sequence that matches the 7-mer seed sequence contained in miR-181a (Figure 5C). Accordingly, elevating miR-181a activity in NMuMG or 4T1 cells reduced Bim protein levels (Figure 5D), while diminishing miR-181a activity elevated the expression of Bim in these same cells (Figure 5E). Moreover, transient transfection of a Bim–3′ UTR–luciferase reporter into NMuMG (Figure 5F) or 4T1 (Figure 5G) cells that stably expressed miR-181a demonstrated that miR-181a did indeed decrease the expression of luciferase containing Bim–3′ UTR. Importantly, mutating the miR-181a seed sequence located within the Bim–3′ UTR prevented miR-181a from suppressing luciferase expression in NMuMG and 4T1 cells (Figure 5, F and G). Thus, miR-181a regulates Bim expression by binding to its complementary seed sequence in the Bim–3′ UTR. It should be noted that Bim mRNA levels were not significantly affected by increasing or decreasing miR-181a activity in these same MECs (Supplemental Figure 8, A–E), indicating that miR-181a regulates cellular levels of Bim by repressing the translation of Bim mRNA, not by inducing its cleavage and degradation. Additionally, pharmacological inhibition of Erk1/2 or Akt failed to alter the ability of miR-181a to decrease Bim protein levels (Supplemental Figure 8, F–H), indicating the miR-181a regulates Bim expression independently of signaling inputs derived from Erk1/2 and Akt. Finally, we attempted to rescue Bim expression by transiently transfecting miR-181a–expressing NMuMG cells with a Bim expression construct that lacked its 3′ UTR sequence, which is targeted by miR-181a. Figure 5H shows that this manipulation not only restored Bim expression to NMuMG cells, but also sensitized them to anoikis and caspase-3 cleavage. Taken together, these findings suggest that miR-181a plays a critical role during the metastatic progression of mammary tumors by downregulating Bim expression, thereby engendering disseminated breast cancer cells with the ability to overcome the physiological barrier imposed by anoikis.
Inhibition of miR-181a abrogates pulmonary tumor outgrowth and increases survival in mice.
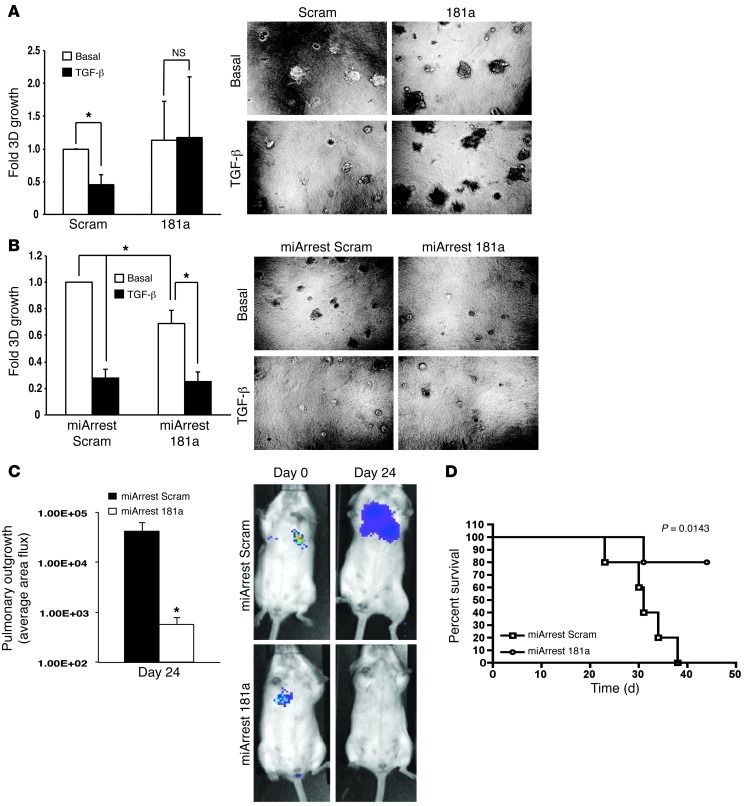
After mammary carcinoma cells escape the primary tumor, circumvent anoikis, traverse the circulation, and invade disseminated organ sites, they ultimately need to reinitiate proliferative programs operant in mediating secondary tumor lesions. As an initial means to investigate the role of miR-181a in regulating metastatic outgrowth, 4T1-luciferase cells engineered to express either miR-181a or a miR-181a sponge (miArrest 181a) were propagated at low densities onto compliant 3D organotypic cultures to recapitulate the pulmonary microenvironment (22, 39–41). Indeed, we recently demonstrated that TGF-β suppresses the outgrowth of 4T1 organoids in compliant 3D culture systems (8). Using bioluminescent growth assays, we now show that elevating miR-181a activity circumvented the cytostatic activities of TGF-β in compliant microenvironments (Figure 6A). Thus, upregulated miR-181a activity may play an additional and important role in overcoming metastatic dormancy. Along these lines, inactivation of miR-181a in 4T1 cells significantly impaired their ability to thrive in 3D pulmonary cultures (Figure 6B), suggesting that measures capable of reducing miR-181a activity may provide a novel therapeutic mechanism to halt the outgrowth of breast cancer micrometastases.
Figure 6 . Inhibition of miR-181a abrogates pulmonary tumor outgrowth and increases survival in mice.
(A and B) The ability of TGF-β1 (5 ng/ml) to suppress 4T1 organoid growth was abrogated by elevated miR-181a activity (A), while basal 4T1 organoid growth was significantly suppressed by miR-181a inactivation (B). Data are the mean ± SEM of bioluminescent signals obtained in 3 independent experiments completed in triplicate. *P < 0.05, Student’s t test. Original magnification, ×20. (C) Luciferase-expressing 4T1 cells engineered to stably express an miR-181a antagonist (i.e., miArrest 181a) were injected into the lateral tail veins of BALB/c mice (n = 5), and pulmonary tumor outgrowth was monitored by intravital bioluminescent imaging. Data are the mean ± SEM of pulmonary photon flux readings 24 days after injection, while the inset shows representative bioluminescent signals of pulmonary outgrowth measured on day 24. *P < 0.05. (D) Kaplan–Meier survival curves of BALB/c mice (n = 5) from C.
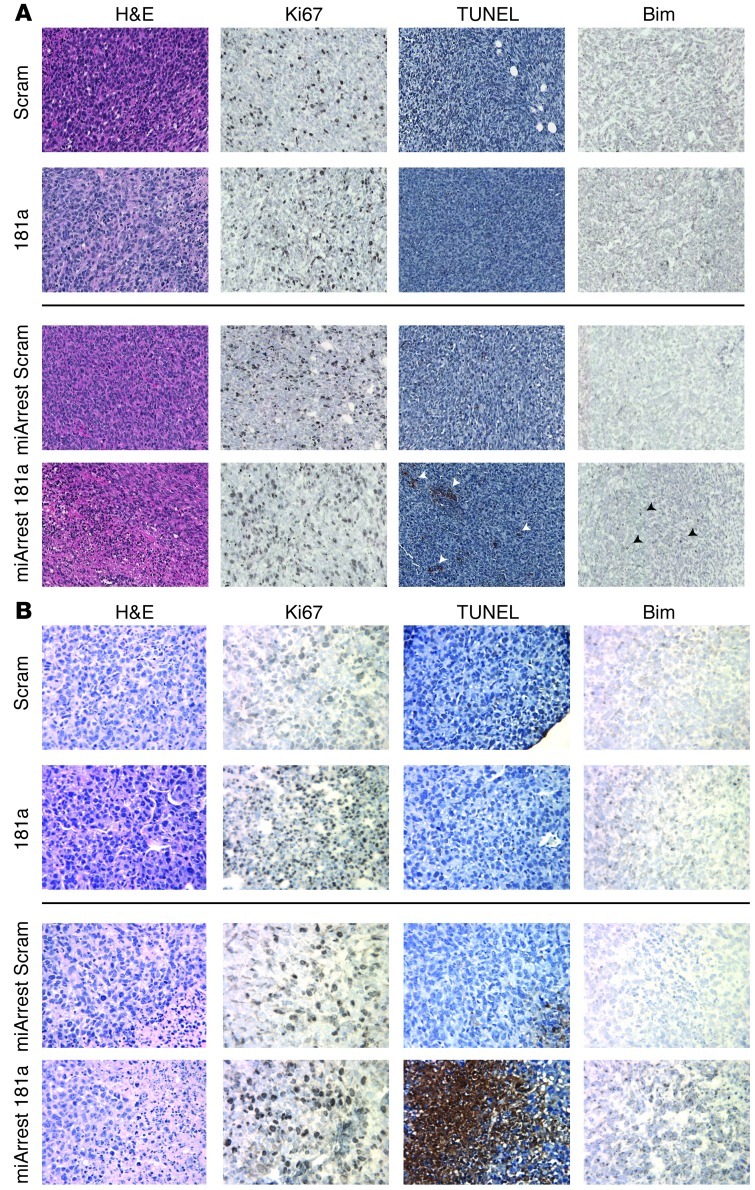
To more rigorously test the above supposition in a preclinical setting, we inoculated the aforementioned 4T1 derivatives into the lateral tail veins of 4-week-old female BALB/c mice. Initial carcinoma cell seeding and subsequent pulmonary outgrowth of 4T1 cells were monitored by biweekly bioluminescent imaging. Inhibiting miR-181a activity decreased pulmonary tumor burden by 4T1 cells (Figure 6C), resulting in significantly increased survival time in mice (Figure 6D). We also investigated the effect of miR-181a expression on 4T1 tumor growth and metastasis from the mammary fat pad. Although manipulation of miR-181a levels failed to significantly alter 4T1 tumor latency, growth, and dissemination from the mammary fat pad (Supplemental Figure 9), we were able to verify that lung metastases that arose from miArrest 181a–expressing 4T1 tumors had in fact lost expression of the miR-181a sponge, as determined by elevated miR-181a biosensor activity in ex vivo cultures of 4T1 lung metastases (data not shown). This result suggests that 4T1 cells that possessed low levels of miR-181a activity were negatively selected during the development and metastasis of 4T1 tumors. Accordingly, immunostaining of 4T1 tumors revealed that those derived from miArrest 181a–expressing 4T1 cells exhibited elevated levels of apoptosis, as measured by increased TUNEL staining and Bim immunoreactivity (Figure 7A). Similar upregulation of TUNEL staining and Bim immunoreactivity were readily detected in the lung metastases produced by miArrest 181a–expressing 4T1 cells, events that were lacking in their parental counterparts (Figure 7B). Collectively, these findings suggest that miR-181a antagonism sensitizes breast cancer cells to undergo enhanced apoptosis via upregulated expression of Bim, an event that may synergize with coadministration of additional DNA damaging agents to induce carcinoma cell death.
Figure 7. Decreased miR-181a expression enhances Bim expression and cell death in murine mammary tumors and metastases.
(A and B) Histopathological analyses of H&E, Ki67, TUNEL, and Bim in 4T1 primary tumors and their pulmonary metastases indicate that inactivation of miR-181a resulted in increased Bim expression and elevated apoptosis. Original magnification, ×20 (A); ×40 (B).
miR-181a is upregulated in TNBCs and corresponds to decreased overall survival times and increased metastasis in breast cancer patients.
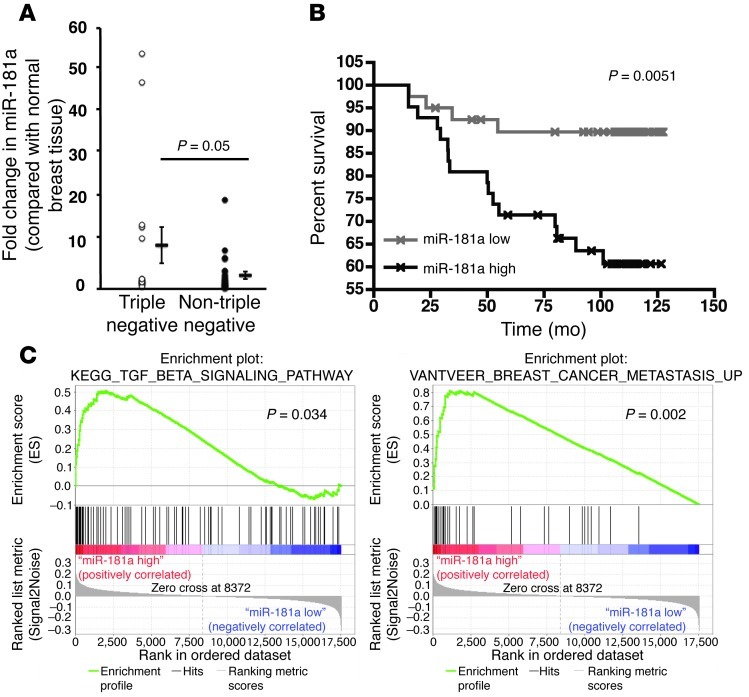
To examine the prognostic value of miR-181a expression in human breast cancers, we performed semiquantitative real-time PCR to monitor miR-181a expression levels in 41 matched cases and adjacent normal tissue samples (Table 1 and ref. 42). In doing so, we determined that TNBCs harbored significantly more miR-181a as compared with non-TNBC subtypes (Figure 8A and Table 1). These findings, together with those presented in Figure 1H, indicate that the upregulated expression of miR-181a is significantly associated with TNBCs relative to other non-TNBC subtypes.
Table 1.
Detection of miR-181a expression levels in human breast cancers
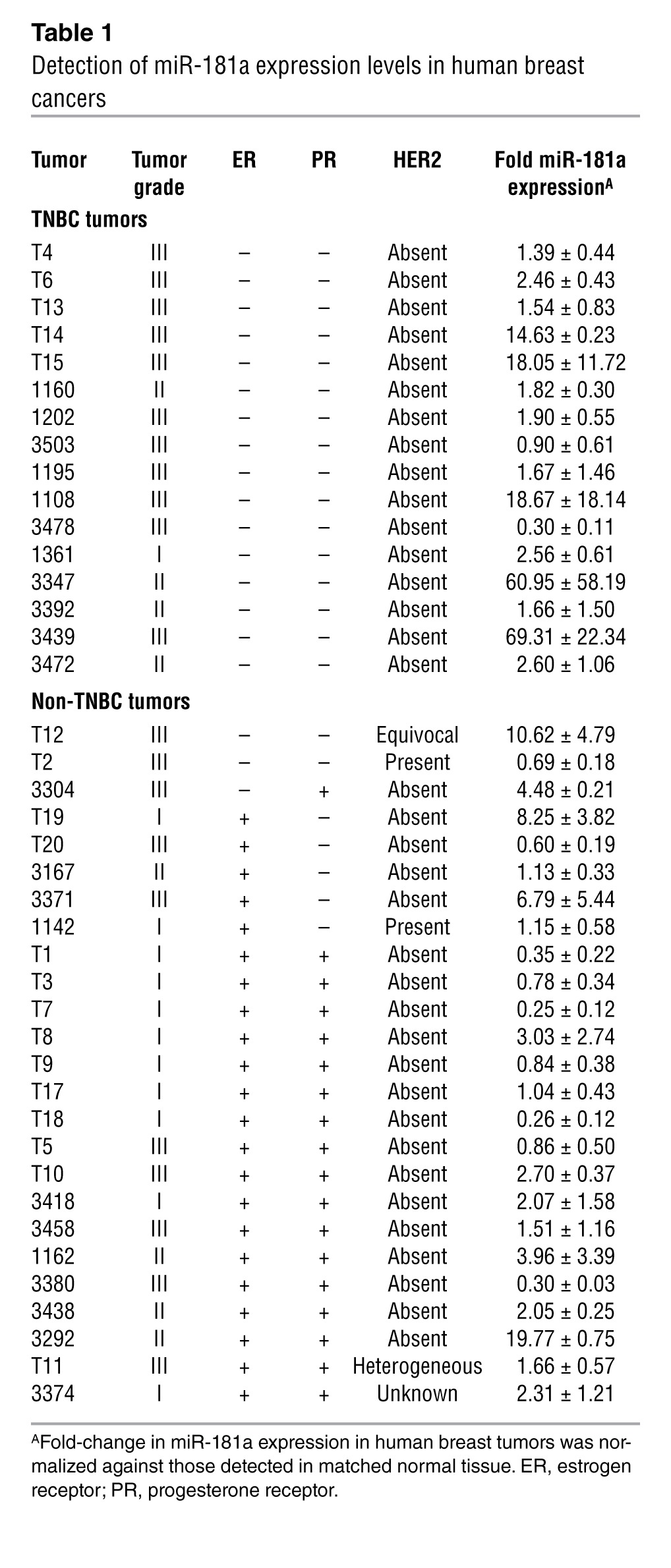
Figure 8. miR-181a is upregulated in TNBCs and corresponds to decreased overall survival times and increased metastasis in breast cancer patients.
(A) Semiquantitative real-time PCR of miR-181a expression in human TNBC (n = 16) as compared with other non-TNBC (n = 25) tumors. Expression levels of miR-181a were normalized to U6 levels and plotted as average fold change detected between the tumor and corresponding normal tissue. Data from 16 TNBC and 25 non-TNBC tumors, displayed as a dot plot alongside the mean ± SEM. P = 0.05. Student’s t test. (B) In an miR expression data set of 82 breast cancer patients that were negative for ErbB2 amplification (43), patients that possessed high tumor expression of miR-181a exhibited significantly reduced disease-free survival times. The median value for miR-181a was used to divide the samples into high (above the median) and low (below the median) miR expression, and the corresponding P value was calculated by log-rank analysis. (C) GSEA (GSE19783) plots for patients with breast tumors that were negative for ErbB2 amplification and expressed high levels of miR-181a demonstrated enrichment for the TGF-β signaling system (44, 45), and for the van’t Veer breast cancer metastasis signature (46).
Finally, we also interrogated a publicly available data set that comprised 101 human primary breast tumor samples that were subjected to genome-wide matched miR and mRNA profiling (43). These analyses showed that the expression of miR-181a significantly predicted for shorter disease-free survival of breast cancer patients whose tumors lacked amplification of the ErbB2 locus (Figure 8B). Moreover, gene set enrichment analysis (GSEA) of the mRNA expression in this same data set clearly showed that mammary tumors harboring high levels of miR-181a expression were significantly enriched for TGF-β signaling (Figure 8C and refs. 44, 45), thereby providing further support for the notion that TGF-β is a master regulator of miR-181a expression in breast cancers. Equally important, we also noted that mammary tumors possessing elevated expression of miR-181a were enriched for the van’t Veer breast cancer metastasis signature (Figure 8C and ref. 46), which comprises a 70-gene signature that forms the basis for the MammaPrint test, which is used clinically to identify breast cancer patients who are at greater risk of developing metastatic disease. Although additional studies are needed to define the role of miR-181a in other specific breast cancer subtypes, our findings nonetheless clearly support an essential function of miR-181a in enhancing breast cancer metastasis and decreasing the long-term survival of breast cancer patients harboring metastatic disease. Collectively, our results implicate miR-181a expression as a potential predictive biomarker for identifying breast cancer patients who are at high risk for developing metastatic disease.
Discussion
It is well established that miRs can function as tumor suppressors (e.g., miR-15a and miR-16-1) and tumor promoters (e.g., miR-155 and miR-21) (47). More recently, miRs have also been implicated in regulating specific steps in the metastatic cascade. Indeed, these “metastamirs” often exhibit little-to-no effect on primary tumor development, but instead can elicit profound effects on either promoting (e.g., miR-10b, miR-143, miR-520c) or suppressing (e.g., miR-31, miR-146a/b, miR-335) metastasis (48, 49). Because metastasis is the major cause of death for breast cancer patients, it stands to reason that defining the molecular mechanisms whereby miRs have an impact on metastasis may provide novel opportunities to treat metastatic breast cancers.
We (8, 22, 50) and others (12, 51) have clearly demonstrated the ability of alterations in the biomechanics of the ECM to have an impact on how cells sense and respond to cellular stimuli, including TGF-β. Here, we significantly expand this theme by demonstrating that changes in ECM rigidity influence coupling of TGF-β to the expression of miRs during breast cancer progression (Figure 1 and Supplemental Tables 1–3). However, it is important to note that individual miRs regulate multiple mRNAs and, likewise, that individual mRNAs are often targeted by multiple miRs. Thus, it is likely that the collective actions of numerous miRs acting on a host of target mRNAs ultimately contribute to the oncogenic functions of TGF-β and its stimulation of breast cancer metastasis. Therefore, future studies need to map the combined actions of miR networks regulated by TGF-β and ECM stiffness and to determine how these events vary between specific breast cancer subtypes and stages of their development.
Along these lines, our current study provides the initial framework to begin these analyses within the context of TNBCs and their acquisition of metastatic phenotypes in response to TGF-β. Indeed, we identified miR-181a as a “metastamir” in TNBCs and demonstrated that measures capable of inhibiting miR-181a activity abrogated TGF-β–mediated EMT, migration, invasion, and metastatic outgrowth in part by upregulating Bim expression. Besides its coupling to Bim expression, miR-181a/b has also been validated to regulate the expression of Bcl-2 (36–38, 52), ATM (15), p27 (17), K-Ras (53), and TIMP3 (16). In the context of TNBCs and the experimental systems used herein, we were unable to associate elevated miR-181a activity with changes in the expression of KLF-6, Smad7, TIMP3, Dusp6, or Bcl-2 (data not shown), suggesting that the manifestations of aberrant miR-181a expression will be determined in a cell- and context-specific manner. Accordingly, recent evidence shows that miR-181a is capable of exhibiting seemingly paradoxical roles in cancer. For example, miR-181a expression is decreased in several cancers, including those of the lung and brain (54). Indeed, elevating miR-181a activity in glioblastomas silences Bcl-2, thereby sensitizing these cells to radiotherapy (36). Thus, in the context of glioblastomas, the expression of miR-181a functions as a tumor suppressor. In stark contrast, aberrantly elevated expression of miR-181a is observed in cancers of the breast (15), mouth (55), liver (16), and blood (56, 57), suggesting that miR-181a fulfills a tumor-promoting role in these contexts. Collectively, these findings and those presented herein highlight the need to identify additional miR-181a targets and define their role in promoting breast cancer development and metastatic progression, particularly that stimulated by TGF-β.
miR-181a belongs to the miR-181 family, which contains 3 other members (i.e., miRs 181b, 181c, and 181d), all of which house identical seed sequences, suggesting that this miR family may exhibit redundancy in targeting mRNAs. The exact mechanism through which TGF-β controls the expression of miR-181 family members remains to be determined. Interestingly, miR-181a and miR-181b are transcribed together from 2 separate genomic loci, while miR-181c and miR-181d are transcribed from a third distinct locus (miRBase; http://www.mirbase.org/). Our findings indicate that Smad4-dependent and -independent mechanisms contribute to TGF-β–mediated transcription of miR-181a; however, miR transcript processing that produces mature miR-181a occurred in a Smad4-independent manner (Supplemental Figure 3, A–C). Thus, future studies need to parse out the relative contributions of transcriptional versus posttranslational mechanisms in mediating TGF-β stimulation of miR-181 family members as well as identify the TGF-β effector molecules operant in mediating these events.
Despite the aforementioned knowledge gaps, our findings do in fact demonstrate that miR-181a plays a unique role in promoting breast cancer metastasis, as the sole inactivation of miR-181a elicited dramatic effects on the ability of breast cancer cells to acquire and maintain EMT, metastatic, and antianoikis phenotypes. Likewise, TGF-β clearly upregulates all miR-181 family members, and therefore, future studies need to determine the specific function and contribution played by miR-181b, miR-181c, and miR-181d during mammary tumorigenesis as well as the extent to which these events are unique or shared among members of this miR family. This point is important because, even though related miRs can exhibit identical seed sequences, they nevertheless can target distinct mRNAs (58). Thus, we suspect that miR-181b, miR-181c, and miR-181d are likely to mediate distinct aspects of mammary tumorigenesis that are different from those currently ascribed to miR-181a.
Evasion of apoptosis is a hallmark of cancer (59), while the evasion of anoikis represents a critical barrier that tumor cells must overcome to metastasize (30). Normally, cell detachment leads to upregulation of Bim, thereby triggering anoikis (30). Our data show that high miR-181a expression repressed that of Bim, thus rendering normal MECs insensitive to anoikis (Figure 5). Conversely, inhibition of miR-181a activity sensitized malignant MECs to anoikis (Figure 5). Interestingly, previous reports have implicated Bim as a critical mediator of chemotherapy-induced apoptosis in numerous cancers (60). For example, Bim expression predicts for an apoptotic response to EGFR inhibitors in EGFR-mutant lung cancers (60). Likewise, high Bim levels are necessary in (a) sensitizing HER2-amplified breast cancer cells to undergo apoptosis in response to lapatinib (60) and (b) mediating breast cancer apoptosis induced by paclitaxel (61). Along these lines, chronic myeloid leukemia patients with low Bim levels have been shown to respond poorly to imatinib (62). Thus, diminished Bim expression not only promotes metastasis, but also supports the development of chemoresistant phenotypes. Our findings suggest that combining miR-181a antagonists with other standard-of-care chemotherapies may provide synergistic benefits by increasing Bim levels, leading to heightened tumor cell sensitivity to apoptosis. Future studies need to address this hypothesis as well as explore the utility of miR-181a as a predictive biomarker for breast cancer metastasis and overall patient survival.
Methods
Cell culture and constructs.
NMuMG, MCF-7, and MDA-MB-231 cells were obtained from ATCC, while isogeneic 67NR, 4T07, and 4T1 cells were provided by Fred Miller (Wayne State University, Detroit, Michigan, USA). MDA-MB-231 cells rendered deficient in Smad4 expression were provided by Yibin Kang (Princeton University, Princeton, New Jersey, USA). Firefly luciferase–expressing 4T1 cells were previously described (63, 64). miRIDIAN miR-181a microRNA Mimics (20 nM final concentration) or Hairpin Inhibitors (50 nM final concentration) were obtained from Thermo Scientific and transiently transfected into cells using Lipofectamine 2000 according to the manufacturer’s recommendations (Life Technologies). NMuMG and 4T1 cells were engineered to possess elevated miR-181a activity by their transduction with miExpress (GeneCopoeia) lentiviral particles that encoded for either control (i.e., scrambled) or precursor miR-181a molecules, followed by the isolation and expansion of polyclonal cell populations by neomycin selection (200 μg/ml; Calbiochem). Alternatively, these same MECs were engineered to possess reduced miR-181a activity by their transduction with miArrest (GeneCopoeia) lentiviral particles that encoded for either control (i.e., scrambled) or miArrest 181a, followed by the isolation and expansion of polyclonal cell populations by hygromycin selection (250 μg/ml; Invitrogen).
miR microarray.
Total RNA was prepared using the QIAGEN RNeasy Plus Mini Kit and following the supplementary protocol for purification of miR (QIAGEN). All experiments were performed in duplicate prior to sending the resulting RNA preparations to Thermo Scientific for miR profiling analyses, whose resulting signal intensities were subjected to statistical filtering that identified miR probes that had P value of 0.05 or less in at least half of the samples. The remaining data were interarray scaled and transformed to log2 values prior to performing 1-way ANOVA analyses for each cell line. Afterward, post hoc analysis was undertaken to identify miRs whose expression was significantly (*P < 0.01) upregulated following administration of TGF-β. The miR expression data are MIAME compliant and have been submitted to GEO (GSE41274).
3D organotypic culture and outgrowth assays.
3D organotypic cultures were performed using the “on-top” method (65) as previously described (8). Briefly, cells were cultured on Cultrex cushions (100%; Trevigen) in complete medium supplemented with 5% Cultrex. ECM rigidity was increased by the addition of type I collagen (3 mg/ml; BD Biosciences) to Cultrex cushions before their solidification. Where indicated, the cells were treated with TGF-β1 (5 ng/ml) for varying lengths of time, and the medium/Cultrex solution was replaced every 4 days. Cells were harvested for RNA analyses by digesting the Cultrex cushions with dispase (BD Biosciences) for 30 minutes at 37°C, followed by inactivation with EDTA (8 mM). Afterward, the dissociated organoids were collected by centrifugation and prepared for immunoblotting or real-time PCR analyses as described below.
Longitudinal bioluminescent growth assays were performed as described (22). Briefly, changes in organoid growth were detected by the addition of D-luciferin (Gold Biotechnology) to induce bioluminescence, which was quantified on a GloMax-Multi Detection System (Promega). Cell proliferation was normalized to initial bioluminescent signals that were obtained 18 hours after plating.
Cell invasion, migration, and proliferation.
Modified Boyden chambers coated with Matrigel (1:25 dilution) were used to monitor the invasiveness of 67NR and 4T1 cells in response to 10% serum and TGF-β1 (5 ng/ml) as described previously (66). Alterations in 4T1 cell migration were assessed by wounding confluent cultures with a micropipette tip, which were immediately placed in serum-free medium supplemented with or without TGF-β1 (5 ng/ml). Bright-field images were obtained immediately after wounding and at various times over a span of 24 hours thereafter. The extent of wound closure was quantitated by measuring the wound areas obtained from 5 independent fields using ImageJ (v1.34s). Finally, alterations in 67NR and 4T1 cell proliferation (10,000 cells/well) were determined by monitoring the incorporation of [3H]thymidine into cellular DNA as described previously (27).
Semiquantitative real-time PCR analyses.
Total RNA was purified using the supplementary miR protocol provided by the RNeasy Plus Mini Kit (QIAGEN). Afterward, cDNAs were synthesized using the miScript Reverse Transcription Kit (QIAGEN) and then diluted 10-fold in H2O prior to their use in semiquantitative real-time PCR reactions that contained 10 μl SsoFast EvaGreen (Bio-Rad), 1 μl miR forward primer, 1 μl miScript Universal Primer (QIAGEN), 3 μl H2O, and 5 μl diluted cDNA. miR expression levels were analyzed on a 7500 Fast Real-Time PCR System (Applied Biosystems). Differences in total RNA concentration were normalized to their corresponding U6 signal. miR precursor levels were determined using the miScript precursor assays as recommended by the manufacturer (QIAGEN). The oligonucleotide primer pairs used are provided in Supplemental Table 4.
Luciferase reporter gene assays.
Normal and malignant MECs were seeded onto 24-well plates (25,000 cells/well) and allowed to adhere overnight. The following morning, the cells were transiently transfected using TransIT-LT1 Transfection Reagent (Mirus) with 50 ng/well of pCMV–β-gal (Clontech) and 450 ng/well psi-Check2 luciferase reporter (Promega) that housed either (a) the complementary seed sequence for miR-181a (e.g., miR-181a biosensor), or (b) the 3′ UTR sequence of Bim that contained either a wild-type or mutant version of the miR-181a seed sequence (wild-type Bim reporter was provided by Clark Distelhorst, Case Western Reserve University). Forty-eight hours after transfection, cells were harvested to measure the quantity of luciferase and β-gal activities present in detergent-solubilized cell extracts. Data are the mean ± SEM luciferase activities of at least 3 independent experiments.
Immunoblotting and immunofluorescence.
Quiescent control and miR-181a–manipulated NMuMG and 4T1 derivatives were incubated in the absence or presence of TGF-β1 (5 ng/ml) for 30 minutes, at which point they were solubilized in Buffer H/1% Triton X-100 and prepared for immunoblotting as described previously (8). Antibodies and pharmacologic inhibitors used herein are described in Supplemental Tables 5 and 6, respectively.
Alterations in the actin cytoskeleton induced by TGF-β in parental and miR-181a–modified cells were monitored using direct TRITC-phalloidin fluorescence as described previously (67).
Anoikis assays.
Control and miR-181a–manipulated NMuMG and 4T1 cells were cultured over poly-HEMA–coated plates in either 0.5% serum or serum-free medium, respectively. The cells were collected at various times over a span of 48 hours, at which point they were prepared for immunoblotting analyses of cleaved caspase-3. For Bim rescue experiments, a rat Bim cDNA that lacked the 3′ UTR was inserted into pcDNA3.1 (Invitrogen) and transiently transfected into control or miR-181a–manipulated cells 24 hours prior to initiating anoikis.
4T1 tumor growth and metastasis assays.
Firefly luciferase–expressing 4T1 cells that stably expressed miR-181a (10,000 cells/injection) or a miR-181a sponge (5,000 cells/injection) were engrafted onto the mammary fat pads of 4-week-old female BALB/c mice. The growth and metastasis of primary 4T1 tumors was quantified by (a) weekly bioluminescent imaging of tumor-bearing animals on a Xenogen IVIS-200 (Caliper Life Sciences) and (b) thrice weekly monitoring of primary tumor size using digital calipers (Fisher). Tumor volumes were calculated by the following equation: tumor volume = 0.5 × x2 × y, where x is the tumor width and y is the tumor length. Primary tumors were excised 4–5 weeks after inoculation, at which point serial histological sections were prepared by the Case Comprehensive Cancer Center’s Tissue Procurement, Histology, & IHC Core, which also performed the H&E and TUNEL staining reactions. Additional immunostaining was undertaken to monitor the expression of Ki-67 (1:50; BD Biosciences — Pharmingen) and Bim (1:50; Cell Signaling) as described previously (68). The resulting images were captured on a D-Metrix DX-40 slide scanner outfitted with Eyepiece Software, or on an Olympus BH2 microscope outfitted with Spot Advanced software (Diagnostic Instruments Inc.).
For pulmonary outgrowth studies, the aforementioned 4T1 derivatives were injected into the lateral tail veins of 4-week-old BALB/c mice. Bioluminescence imaging was performed 30 minutes after inoculation (T0) and biweekly thereafter. Pulmonary tumor development was assessed by normalizing biweekly images to those obtained at T0.
Analysis of human miR and mRNA microarray data.
Expression and clinical outcome data were obtained from a publicly available data set of 101 human primary breast tumor samples, which contained matched genome-wide miR and mRNA profiling (GSE19783; ref. 43). All samples were median-centered for miR-181a expression and denoted as having high expression if individual miR-181a signals fell above the median and as having low expression if individual miR-181a signals fell below the median. Kaplan-Meier curves were generated using GraphPad Prism for Macintosh (v5.0). mRNA expression profiles for tumors not classified as being ErbB2 amplified were divided into high (n = 41) or low (n = 41) expression of miR-181a, and GSEA was performed using GSEA software (v2.07) that was obtained from the Broad Institute. Genes were analyzed for enrichment using the c2 gene sets from the MSigDB, which contains 3271 curated gene sets obtained from public pathway databases and published gene signatures. P values, enrichment scores (ES), and q values were computed by permuting the sample labels (phenotype) 1,000 times (69, 70).
Study approval.
All animal procedures were performed in accordance to protocols approved by the Institutional Animal Care and Use Committee for Case Western Reserve University. For human breast cancer specimens, primary tumors and matched normal tissues were collected and processed under approved IRB protocols from the University Hospitals Case Medical Center and the Cleveland Clinic. All patients provided written informed consent and consented to allowing the study investigators access to their tumor specimens and clinical data.
Statistics.
Statistical values were defined using a 2-tailed unpaired Student’s t test, where P < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank members of the Schiemann Laboratory for critical comments and reading of the manuscript. We also thank Anna L. Smith and Heide L. Ford for helpful discussions related to miR-181a data mining techniques, and Clark W. Distelhorst for providing the Bim–3′ UTR luciferase plasmid. W.P. Schiemann was supported by grants from the NIH (CA129359), the Komen Foundation (BCTR0706967), and the University of Colorado Cancer Center Breast Program Gift Fund. Additional support to W.P. Schiemann and K. Sossey-Alaoui was provided by pilot funds from the Case Comprehensive Cancer Center (P30 CA043703), which also provided expertise through the Athymic Animal and Xenograft Core, the Imaging Research Core, and the Tissue Procurement, Histology, & IHC core. Finally, M.A. Taylor was supported by the Department of Defense (BC093128).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2013;123(1):150–163. doi:10.1172/JCI64946.
References
- 1.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147(2):275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 3.Taylor MA, Parvani JG, Schiemann WP. The pathophysiology of epithelial-mesenchymal transition induced by transforming growth factor-β in normal and malignant mammary epithelial cells. J Mammary Gland Biol Neoplasia. 2010;15(2):169–190. doi: 10.1007/s10911-010-9181-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parvani JG, Taylor MA, Schiemann WP. Noncanonical TGF-β signaling during mammary tumorigenesis. J Mammary Gland Biol Neoplasia. 2011;16(2):127–146. doi: 10.1007/s10911-011-9207-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang YE. Non-Smad pathways in TGF-β signaling. Cell Res. 2009;19(1):128–139. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levental KR, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139(5):891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lopez JI, Kang I, You WK, McDonald DM, Weaver VM. In situ force mapping of mammary gland transformation. Integr Biol (Camb). 2011;3(9):910–921. doi: 10.1039/c1ib00043h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor MA, Amin JD, Kirschmann DA, Schiemann WP. Lysyl oxidase contributes to mechanotransduction-mediated regulation of transforming growth factor-β signaling in breast cancer cells. Neoplasia. 2011;13(5):406–418. doi: 10.1593/neo.101086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elenbaas B, Weinberg RA. Heterotypic signaling between epithelial tumor cells and fibroblasts in carcinoma formation. Exp Cell Res. 2001;264(1):169–184. doi: 10.1006/excr.2000.5133. [DOI] [PubMed] [Google Scholar]
- 10.Erler JT, et al. Hypoxia-induced lysyl oxidase is a critical mediator of bone marrow cell recruitment to form the premetastatic niche. Cancer Cell. 2009;15(1):35–44. doi: 10.1016/j.ccr.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erler JT, et al. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature. 2006;440(7088):1222–1226. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- 12.Erler JT, Weaver VM. Three-dimensional context regulation of metastasis. Clin Exp Metastasis. 2009;26(1):35–49. doi: 10.1007/s10585-008-9209-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butcher DT, Alliston T, Weaver VM. A tense situation: forcing tumour progression. Nat Rev Cancer. 2009;9(2):108–122. doi: 10.1038/nrc2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11(3):228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, et al. Transforming growth factor-β regulates the sphere-initiating stem cell-like feature in breast cancer through miRNA-181 and ATM. Oncogene. 2011;30(12):1470–1480. doi: 10.1038/onc.2010.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang B, et al. TGFβ-mediated upregulation of hepatic miR-181b promotes hepatocarcinogenesis by targeting TIMP3. Oncogene. 2010;29(12):1787–1797. doi: 10.1038/onc.2009.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang B, Li W, Guo K, Xiao Y, Wang Y, Fan J. miR-181b Promotes hepatic stellate cells proliferation by targeting p27 and is elevated in the serum of cirrhosis patients. Biochem Biophys Res Commun. 2012;421(1):4–8. doi: 10.1016/j.bbrc.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 18.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454(7200):56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong W, et al. MicroRNA-155 is regulated by the transforming growth factor β/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol Cell Biol. 2008;28(22):6773–6784. doi: 10.1128/MCB.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blahna MT, Hata A. Smad-mediated regulation of microRNA biosynthesis. FEBS Lett. 2012;586(14):1906–1912. doi: 10.1016/j.febslet.2012.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paszek MJ, Weaver VM. The tension mounts: mechanics meets morphogenesis and malignancy. J Mammary Gland Biol Neoplasia. 2004;9(4):325–342. doi: 10.1007/s10911-004-1404-x. [DOI] [PubMed] [Google Scholar]
- 22.Wendt MK, Taylor MA, Schiemann BJ, Schiemann WP. Down-regulation of epithelial cadherin is required to initiate metastatic outgrowth of breast cancer. Mol Biol Cell. 2011;22(14):2423–2435. doi: 10.1091/mbc.E11-04-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992;52(6):1399–1405. [PubMed] [Google Scholar]
- 24.Micalizzi DS, et al. The Six1 homeoprotein induces human mammary carcinoma cells to undergo epithelial-mesenchymal transition and metastasis in mice through increasing TGF-β signaling. J Clin Invest. 2009;119(9):2678–2690. doi: 10.1172/JCI37815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maillot G, et al. Widespread estrogen-dependent repression of micrornas involved in breast tumor cell growth. Cancer Res. 2009;69(21):8332–8340. doi: 10.1158/0008-5472.CAN-09-2206. [DOI] [PubMed] [Google Scholar]
- 26.Neil JR, Schiemann WP. Altered TAB1:IκB kinase interaction promotes transforming growth factor β-mediated nuclear factor-κB activation during breast cancer progression. Cancer Res. 2008;68(5):1462–1470. doi: 10.1158/0008-5472.CAN-07-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galliher AJ, Schiemann WP. β3 integrin and Src facilitate transforming growth factor-β mediated induction of epithelial-mesenchymal transition in mammary epithelial cells. Breast Cancer Res. 2006;8(4):R42. doi: 10.1186/bcr1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miettinen PJ, Ebner R, Lopez AR, Derynck R. TGF-β induced transdifferentiation of mammary epithelial cells to mesenchymal cells: involvement of type I receptors. J Cell Biol. 1994;127(6 pt 2):2021–2036. doi: 10.1083/jcb.127.6.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature. 2003;425(6958):577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 30.Nagaprashantha LD, Vatsyayan R, Lelsani PC, Awasthi S, Singhal SS. The sensors and regulators of cell-matrix surveillance in anoikis resistance of tumors. Int J Cancer. 2011;128(4):743–752. doi: 10.1002/ijc.25725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoshino Y, Katsuno Y, Ehata S, Miyazono K. Autocrine TGF-β protects breast cancer cells from apoptosis through reduction of BH3-only protein, Bim. J Biochem. 2011;149(1):55–65. doi: 10.1093/jb/mvq114. [DOI] [PubMed] [Google Scholar]
- 32.Neil JR, Tian M, Schiemann WP. x-linked inhibitor of apoptosis protein and its E3 ligase activity promote transforming growth factor-β-mediated nuclear factor-κB activation during breast cancer progression. J Biol Chem. 2009;284(32):21209–21217. doi: 10.1074/jbc.M109.018374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27(1):91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2(11):e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akiyama T, Tanaka S. Bim: guardian of tissue homeostasis and critical regulator of the immune system, tumorigenesis and bone biology. Arch Immunol Ther Exp. 2011;59(4):277–287. doi: 10.1007/s00005-011-0126-1. [DOI] [PubMed] [Google Scholar]
- 36.Chen G, et al. MicroRNA-181a sensitizes human malignant glioma U87MG cells to radiation by targeting Bcl-2. Oncol Rep. 2010;23(4):997–1003. doi: 10.3892/or_00000725. [DOI] [PubMed] [Google Scholar]
- 37.Li H, Hui L, Xu W. miR-181a sensitizes a multidrug-resistant leukemia cell line K562/A02 to daunorubicin by targeting BCL-2. Acta Biochim Biophys Sin. 2012;44(3):269–277. doi: 10.1093/abbs/gmr128. [DOI] [PubMed] [Google Scholar]
- 38.Ouyang YB, Lu Y, Yue S, Giffard RG. miR-181 targets multiple Bcl-2 family members and influences apoptosis and mitochondrial function in astrocytes. Mitochondrion. 2012;12(2):213–219. doi: 10.1016/j.mito.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barkan D, et al. Metastatic growth from dormant cells induced by a col-I-enriched fibrotic environment. Cancer Res. 2010;70(14):5706–5716. doi: 10.1158/0008-5472.CAN-09-2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barkan D, et al. Inhibition of metastatic outgrowth from single dormant tumor cells by targeting the cytoskeleton. Cancer Res. 2008;68(15):6241–6250. doi: 10.1158/0008-5472.CAN-07-6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shibue T, Weinberg RA. Integrin β1-focal adhesion kinase signaling directs the proliferation of metastatic cancer cells disseminated in the lungs. Proc Natl Acad Sci U S A. 2009;106:10290–10295. doi: 10.1073/pnas.0904227106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sossey-Alaoui K, Downs-Kelly E, Das M, Izem L, Tubbs R, Plow EF. WAVE3, an actin remodeling protein, is regulated by the metastasis suppressor microRNA, miR-31, during the invasion-metastasis cascade. Int J Cancer. 2011;129(6):1331–1343. doi: 10.1002/ijc.25793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Enerly E, et al. miRNA-mRNA integrated analysis reveals roles for miRNAs in primary breast tumors. PLoS One. 2011;6(2):e16915. doi: 10.1371/journal.pone.0016915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40(Database issue):D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van ‘t Veer LJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 47.Kent OA, Mendell JT. A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene. 2006;25(46):6188–6196. doi: 10.1038/sj.onc.1209913. [DOI] [PubMed] [Google Scholar]
- 48.Dykxhoorn DM. MicroRNAs and metastasis: little RNAs go a long way. Cancer Res. 2010;70(16):6401–6406. doi: 10.1158/0008-5472.CAN-10-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hurst DR, Edmonds MD, Welch DR. Metastamir: the field of metastasis-regulatory microRNA is spreading. Cancer Res. 2009;69(19):7495–7498. doi: 10.1158/0008-5472.CAN-09-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Allington TM, Galliher-Beckley AJ, Schiemann WP. Activated Abl kinase inhibits oncogenic transforming growth factor-β signaling and tumorigenesis in mammary tumors. FASEB J. 2009;23(12):4231–4243. doi: 10.1096/fj.09-138412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leight JL, Wozniak MA, Chen S, Lynch ML, Chen CS. Matrix rigidity regulates a switch between TGF-β1-induced apoptosis and epithelial-mesenchymal transition. Mol Biol Cell. 2012;23(5):781–791. doi: 10.1091/mbc.E11-06-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu W, Shan X, Wang T, Shu Y, Liu P. miR-181b modulates multidrug resistance by targeting BCL2 in human cancer cell lines. Int J Cancer. 2010;127(11):2520–2529. doi: 10.1002/ijc.25260. [DOI] [PubMed] [Google Scholar]
- 53.Shin KH, Bae SD, Hong HS, Kim RH, Kang MK, Park NH. miR-181a shows tumor suppressive effect against oral squamous cell carcinoma cells by downregulating K-ras. Biochem Biophys Res Commun. 2010;404(4):896–902. doi: 10.1016/j.bbrc.2010.12.055. [DOI] [PubMed] [Google Scholar]
- 54.Seoudi AM, Lashine YA, Abdelaziz AI. MicroRNA-181a — a tale of discrepancies. Expert Rev Mol Med. 2012;14:e5. doi: 10.1017/S1462399411002122. [DOI] [PubMed] [Google Scholar]
- 55.Yang CC, et al. miR-181 as a putative biomarker for lymph-node metastasis of oral squamous cell carcinoma. J Oral Pathol Med. 2011;40(5):397–404. doi: 10.1111/j.1600-0714.2010.01003.x. [DOI] [PubMed] [Google Scholar]
- 56.Pons A, et al. Hematopoiesis-related microRNA expression in myelodysplastic syndromes. Leuk Lymphoma. 2009;50(11):1854–1859. doi: 10.3109/10428190903147645. [DOI] [PubMed] [Google Scholar]
- 57.Pichiorri F, et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc Natl Acad Sci U S A. 2008;105(35):12885–12890. doi: 10.1073/pnas.0806202105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133(2):217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 60.Faber AC, et al. BIM expression in treatment naive cancers predicts responsiveness to kinase inhibitors. Cancer Discov. 2011;1(4):352–365. doi: 10.1158/2159-8290.CD-11-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sunters A, et al. FoxO3a transcriptional regulation of Bim controls apoptosis in paclitaxel-treated breast cancer cell lines. J Biol Chem. 2003;278(50):49795–49805. doi: 10.1074/jbc.M309523200. [DOI] [PubMed] [Google Scholar]
- 62.San Jose-Eneriz E, et al. Epigenetic down-regulation of BIM expression is associated with reduced optimal responses to Imatinib treatment in chronic myeloid leukaemia. Eur J Cancer. 2009;45(10):1877–1889. doi: 10.1016/j.ejca.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 63.Wendt MK, Smith JA, Schiemann WP. p130Cas is required for mammary tumor growth and TGF-β-mediated metastasis through regulation of Smad2/3 activity. J Biol Chem. 2009;284(49):34145–31156. doi: 10.1074/jbc.M109.023614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wendt MK, Cooper AN, Dwinell MB. Epigenetic silencing of CXCL12 increases the metastatic potential of mammary carcinoma cells. Oncogene. 2008;27(10):1461–1471. doi: 10.1038/sj.onc.1210751. [DOI] [PubMed] [Google Scholar]
- 65.Lee GY, Kenny PA, Lee EH, Bissell MJ. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods. 2007;4(4):359–365. doi: 10.1038/nmeth1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schiemann WP, Blobe GC, Kalume DE, Pandey A, Lodish HF. Context-specific effects of fibulin-5 (DANCE/EVEC) on cell proliferation, motility, and invasion. Fibulin-5 is induced by transforming growth factor-β and affects protein kinase cascades. J Biol Chem. 2002;277(30):27367–27377. doi: 10.1074/jbc.M200148200. [DOI] [PubMed] [Google Scholar]
- 67.Sokol JP, Neil JR, Schiemann BJ, Schiemann WP. The use of cystatin C to inhibit epithelial-mesenchymal transition and morphological transformation stimulated by transforming growth factor-β. Breast Cancer Res. 2005;7(5):R844–R853. doi: 10.1186/bcr1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Galliher-Beckley AJ, Schiemann WP. Grb2 binding to Tyr284 in TbetaR-II is essential for mammary tumor growth and metastasis stimulated by TGF-beta. Carcinogenesis. 2008;29(2):244–251. doi: 10.1093/carcin/bgm245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.