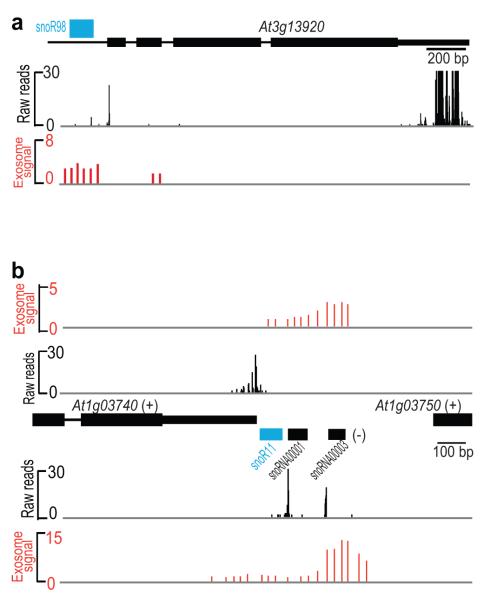
Figure 4. Identification of ncRNAs at Sites Affected by the Exosome.
(a) Comparison of DRS reads (black) with exosome knockdown array data (red) for the At3g13920 open reading frame, suggested to be regulated by the exosome24, which also encodes a known but un-annotated snoRNA (blue). (b) Comparison of DRS reads (black) with exosome knockdown array data (red) showing differences in strand specificity. The previously proposed role of the exosome in controlling 3′ end formation at At1g03740 may now be explained by processing of snoRNAs that are not annotated (blue) or annotated (black) in TAIR10 on the other strand and artifacts resulting from reverse transcriptase and tiling arrays.