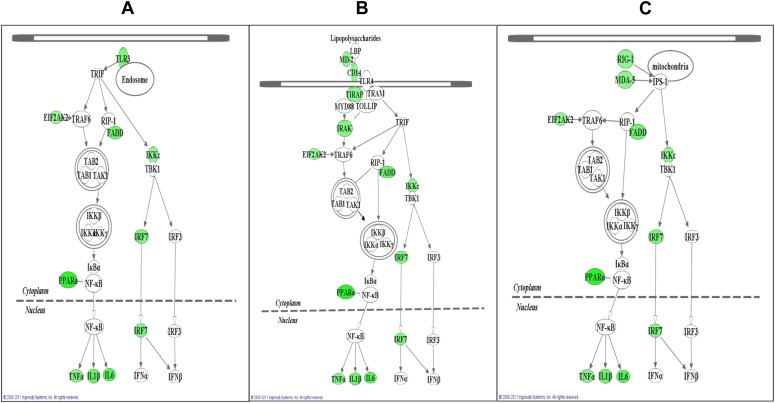
Fig. 1.
Genes whose mRNA expression is suppressed by nicotine in RAW264.7 cells under poly(I:C) stimulation compared with those under poly(I:C) stimulation only are mapped to three gene networks, as predicted by ingenuity pathway analysis (IPA). Cells were stimulated with poly(I:C) at 20 μg/ml with and without prior treatment with 5 μM nicotine and then subjected to quantitative RT-PCR analysis (qRT-PCR) using an array. A total of 51 genes was assayed, including two control genes (β-actin and GAPDH) (Table 1). These genes could be classified into three pathways following IPA analysis: (A) TLR3, which is a major receptor for poly(I:C), depends on the TRIF pathway, and leads to activation of IRFs and NF-κB; (B) TLR4, which depends on both the TRIF and MyD88 pathways and mediates bacteria-induced inflammation; and (C) RLR, whose receptor mRNA expression can be induced by TLR3 and that shares similar downstream signaling with TLR3 and plays a key role in virus-induced innate immune responses. mRNA expression of 15 genes was found to be significantly suppressed in RAW264.7 cells when nicotine-treated cells were further challenged with poly(I:C) (Table 1) compared with the poly(I:C)-only treated cells (P < 0.05). These significantly downregulated genes are highlighted in green in the three pathways described above. For a detailed summary of expression change of each gene, please see Table 2.