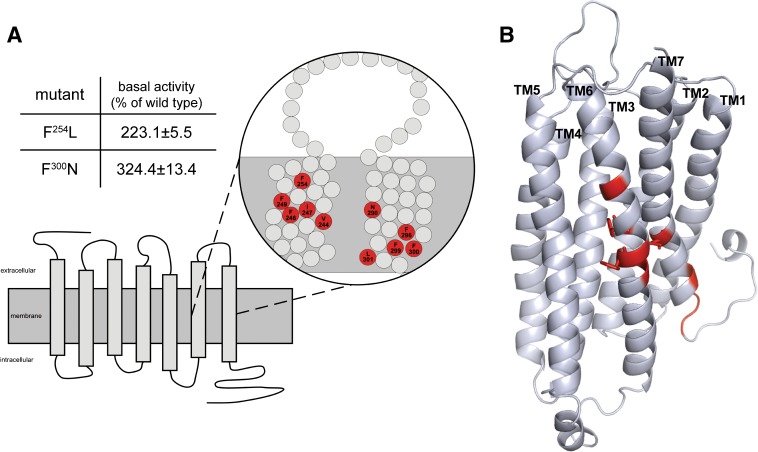
Fig. 4.
Position and basal activity of constitutively active P2Y12 mutants. (A) the position of constitutively active mutations in transmembranes 1 and 7 (TM6 and TM7) are depicted. Basal activities of the individual mutants expressed in yeast are given in the table. Data are presented as mean ± S.D. of three independent experiments, each carried out in triplicate. The basal activity of the WT P2Y12 was OD600 nm: 0.074±0.016. Complete functional data are available and organized in a P2Y12 mutant database (http://www.ssfa-7tmr.de/p2y12). (B) the comparative model of P2Y12 based on the CXCR4 template is depicted. Residues producing constitutively active mutants on TM6 and TM7 are highlighted in red. Residue side chains facing the pore of the receptor (F246, F249, and N290) are shown in sticks.