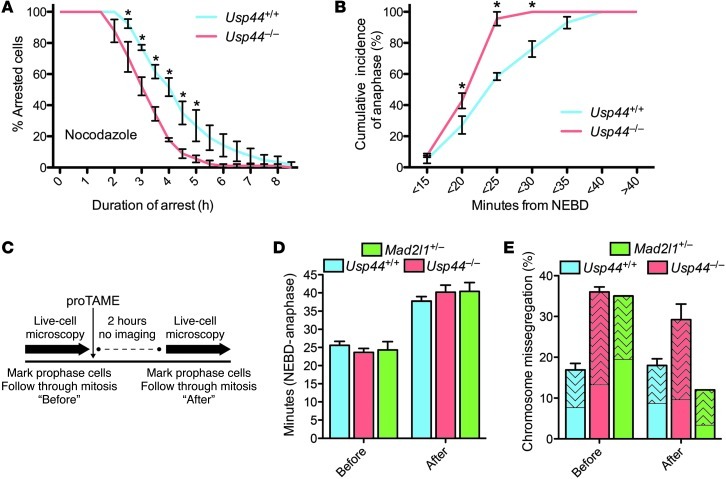
Figure 3. Correction of the checkpoint defect in Usp44-null cells does not prevent mitotic errors.
(A) MEFs of the indicated genotype were transduced with H2B-YFP, followed by live-cell microscopy. Mitotic arrest duration was determined by calculating the time from nuclear envelope breakdown (NEBD) to chromosome decondensation. Graph represents the mean percentage ± SEM of cells remaining in mitosis at each time point (3 lines per genotype). Total n = 50–90 cells for each genotype per condition. (B) The interval between NEBD and anaphase was determined in the absence of nocodazole for MEFs of the indicated genotypes. The graph represents the cumulative incidence of anaphase at each time point (mean ± SEM of 3 lines). Total n = 50–60 cells for each genotype. (C) Experimental scheme for D and E. MEFs expressing H2B-YFP were marked in prophase and followed by time-lapse microscopy to determine mitotic duration and chromosome mis-segregation. Two hours after the addition of proTAME, new prophase cells were selected and similarly examined. (D) Duration of mitosis for Usp44+/+, Usp44–/–, and Mad2l1+/– MEFs imaged before and after proTAME addition. The graph represents the mean ± SEM for 3 lines per genotype. Total n = 25–35 cells per time period. (E) The effect of proTAME on chromosome mis-segregation in Usp44+/+ (n = 67 before, 83 after), Usp44–/– (n = 65 cells per time period from 3 MEF lines), and Mad2l1+/– MEFs (n = 20–25 cells per time period, 1 MEF line). The patterned area indicates the proportion of cells encountering at least one lagging chromosome. The graph represents the mean ± SEM. *P < 0.05, 2-way ANOVA.