Figure 5.
Mechanisms for Demethylation
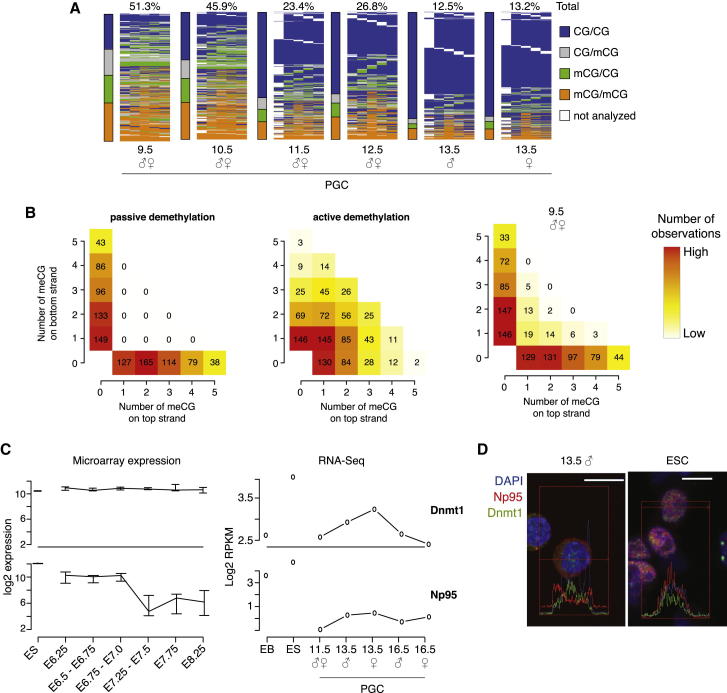
(A) Hairpin bisulfite heatmap of LINE1Tf. Total methylation levels are shown at the top. For each time point analyzed, each column represents one CG dyad along the LINE1Tf consensus sequence, and each row represents one sequencing read. The bars next to the heatmap represent the average distribution of fully methylated, hemimethylated, and unmethylated sites. Note that E9.5 and E10.5 PGCs have high levels of hemimethylated sites, which are then reduced to almost-complete hypomethylation at E13.5.
(B) Shown is the distribution of methylated CG dinucleotides (meCG) at hemimethylated sites across the top (x axis) and bottom (y axis) strands of the LINE1Tf consensus sequence assessed by hairpin bisulfite sequencing. The LINE1Tf consensus sequence contains five CG dinucleotides, and the numbers 0–5 on the axis refer to the amount of meCGs on each strand and contain no position information. The values in the heat diagram represent the number of instances with the respective number of meCGs observed on the top and bottom strands. Shown is a simulation of the distribution of meCG within hemimethylated sites in the case of passive DNA demethylation (left) and active demethylation (middle). Note that with passive demethylation, all meCGs are located on the top strand, while the bottom strand is completely unmethylated and contains 0 meCGs and vice versa. For active DNA demethylation, a strand-independent distribution was simulated that leads to methylated and unmethylated CGs randomly distributed across both strands. The hairpin bisulfite data for E9.5 PGCs are shown in the right panel, and there is a strong strand bias for meCGs toward either top or bottom strand highly similar to the outcome for the simulation of passive DNA demethylation. Instances with meCGs distributed across both strands are rare in E9.5 PGCs. See also Figure S6.
(C) Expression analysis of the DNA methylation machinery. Single-cell microarray data for ESCs (Vincent et al., 2011) and PGCs (Kurimoto et al., 2008) were reanalyzed (left, see the Experimental Procedures for more detail). RNA-Seq data for ESC and embryoid body (EB) (Cloonan et al., 2008) and PGCs of various time points are shown on the right. Whiskers represent the interquartile range of variation between replicates. Note that while Dnmt1 is continuously expressed, Np95 is transcriptionally downregulated in early PGCs. See also Figure S7.
(D) Immunofluorescence staining for DNA (blue), Dnmt1 (green), and Np95 (red). Shown are immunostainings and RGB profiles created with Zeiss LSM software. Scale bars represent 10 μM in all images, and where RGB profiles are shown, the red line across a cell represents the midline along which the signal intensity is traced for each pixel and the profile is plotted below. Shown are stainings for Np95 and Dnmt1 in E13.5 male PGCs and E14 ESCs. In cycling PGCs, Dnmt1 localizes to the nucleus while Np95 is preferentially located in the cytoplasm. In ESCs, both Dnmt1 and Np95 localize to the nucleus. This suggests that in ESCs, the subcellular localization of Dnmt1 and Np95 is linked during S phase, while this dynamic pattern may be uncoupled in PGCs. See also Figure S7.