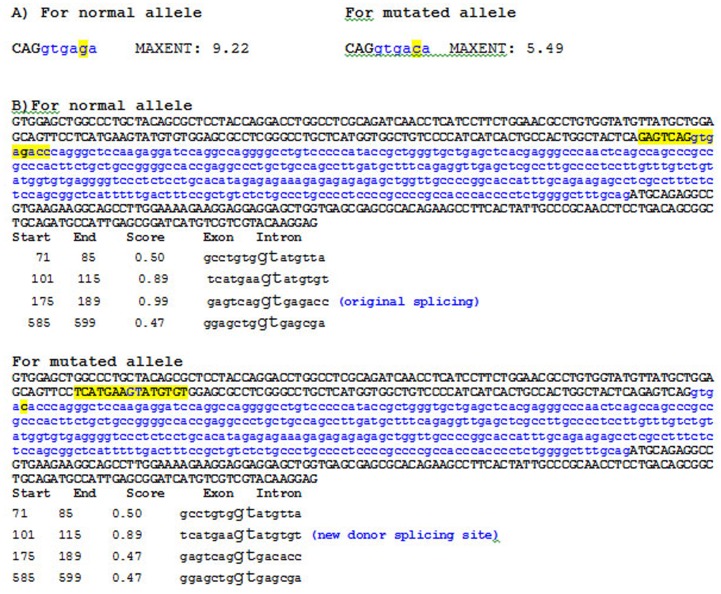
Figure 7. Analysis of the c.1081+5G<T intronic mutation using ME and NN programs.
A) Numerical result produced by the ME program. Three nucleotides in uppercase correspond to exon 2, in blue intron 2. The normal and changed alleles in position +5 (c.1081+5G>C) are highlighted in yellow. The predicted number for the mutant allele falls four points, which could alter the entropy. B) Analysis of c.1081+5G<T intronic mutation with NN program. Exon 2 (181 nucleotides), intron 2 (360) and exon 3 (143) were represented. The nucleotides in uppercase correspond to exon 2 and exon 3, in blue intron 2. For the normal allele: the site of splice original is highlighted in yellow. For the mutated allele: the change from g to c seems to produce the loss of the normal recognition splice site and there is an 89% chance to recognize a cryptic donor splice site located within exon 2. This sequence is highlighted in yellow.