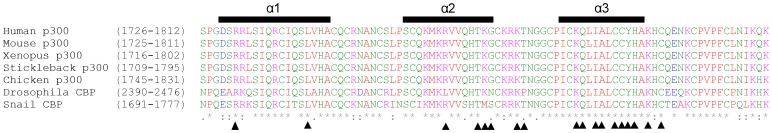
Figure 8. Multiple sequence alignment of the highly homologous TAZ2 domains of p300 and CBP.
The multiple sequence alignment of the TAZ2 domain of human, mouse, western clawed frog (Xenopus tropicalis), stickleback and chicken p300, and drosophila and pond snail CBP, illustrates the high degree of sequence homology between the TAZ2 domains of a diverse range of species. Residues are coloured according to the residue type, with small and hydrophobic residues in red (AVFPMILW), acidic residues in blue (DE), basic residues in magenta (RK) and residues containing a hydroxyl, sulfhydryl or sidechain amide group in green (STYHCNQ). Glycine was also coloured in green. Consensus symbols are shown below the sequence. Residues marked with an ‘*’ were fully conserved between sequences. The symbol ‘:’ indicates conservation between groups with strongly similar properties and ‘.’ indicates conservation between groups of weakly similar properties. TAZ2 residues that were significantly shifted upon binding to B-Myb are indicated by triangles shown below the consensus. The positions of the helices in p300 TAZ2, which were identified by analysis of the backbone resonance assignments using the chemical shift index method are indicated above the sequence. The alignment was prepared using ClustalW.