Abstract
Determination of hepatocyte proliferation activity is hampered by the presence of Ki67-positive non-parenchymal cells. We validated a multicolor immunohistochemical (IHC) approach using multispectral tissue and cell segmentation software. Portal vein branches to the cranial liver lobes of 10 rabbits were embolized, leading to atrophy of the cranial lobes and hyperplasia of the caudal lobes. Slides from cranial and caudal lobes (n=20) were double-stained (CK8+18 and Ki67) and triple-stained (CK8+18, Ki67, and CD31). The Ki67 proliferation index was calculated using automated tissue and cell segmentation software and compared with manual counting by two independent observers. A substantial variation was seen in the number of Ki67-positive hepatocytes in the different specimens in both double and triple staining (range, 0–50). Correlation coefficients between manual counting and the digital analysis were 0.76 for observer 1 (p<0.001) and 0.78 for observer 2 (p<0.001) with double staining and R2 = 0.91 for observer 1 and R2 = 0.89 for observer 2, p<0.001 with triple staining. In conclusion, in rabbit, the hepatocellular proliferation index can be reliably determined using automated tissue and cell segmentation software in combination with IHC multiple staining. Our findings may be useful in clinical practice when Ki67 proliferation index yields prognostic significance.
Keywords: immunohistochemistry, multiple immunostaining, rabbit liver, hepatocytes, proliferation, automated scoring, quantitation, segmentation
Liver resection is the most effective treatment for primary or metastatic liver tumors. The extent of liver resection is restricted by the minimum volume of liver remnant required to provide sufficient postoperative liver function. Preoperative portal vein embolization (PVE) is an option to increase future remnant liver (FRL) volume through induction of regeneration of the hepatocellular mass of the FRL (de Graaf et al. 1998). Following occlusion of the right or left branch of the portal vein, atrophy of the embolized liver segments occurs, whereas hyperplasia of the contralateral, non-embolized liver lobe is induced. In our group, we study the effects of PVE of the liver in different circumstances in a rabbit model as developed by de Graaf et al. (2011; van den Esschert et al. 2011). One of the parameters of interest in this model is the Ki67 proliferation index in both the embolized and non-embolized liver lobes in different research settings. However, one of the difficulties we encountered was the presence of non-parenchymal cells, such as leucocytes in the sinusoidal space of the liver, which stain positive for Ki67 and interfere with reliable determination of the proliferation of hepatocytes (Fig. 1). Particularly in the cranial embolized lobe with low proliferation activity of the hepatocytes, there may be a significant influx of inflammatory cells (van den Esschert et al. 2012).
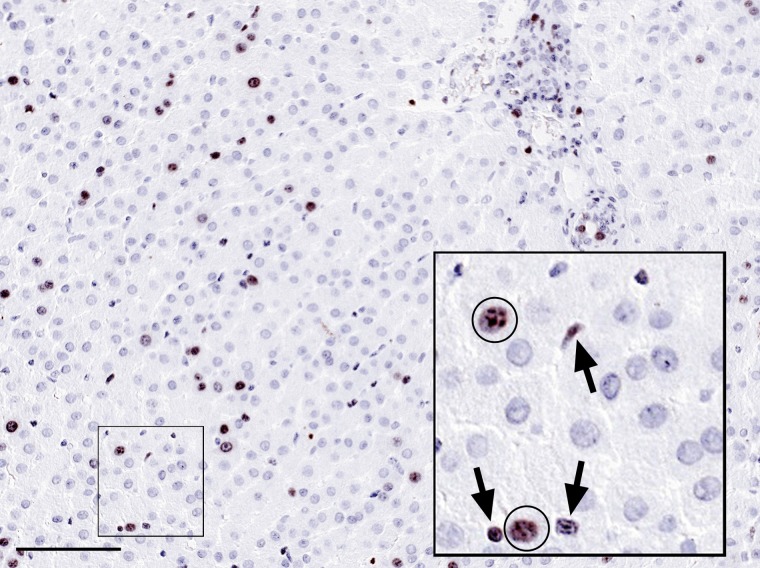
Figure 1.
Formalin-fixed and paraffin-embedded rabbit liver tissue section showing the presence of a substantial number of Ki67-positive nuclei in brown and weakly counterstained with hematoxylin. It is mostly unclear if these cells are proliferating hepatocytes or non-parenchymal cells, of which sinusoidal lymphocytes may be particularly difficult to distinguish from hepatocytes, both having round nuclei. Insert shows a high magnification with Ki67-positive nuclei from hepatocytes (circles), whereas the nuclei depicted by arrows are non-parenchymal cells in the sinusoidal space, including lymphocytes. Bar = 0.1 mm.
Multispectral imaging (MSI) has been applied for unmixing double, triple, and quadruple immunohistochemistry (IHC) plus hematoxylin counterstain based on the individual spectral characteristics of chromogens and dyes (van der Loos 2008, 2010). MSI allows quantitation of reaction products in the individual component images, as well as visualization and quantitation of co-localization (Levenson 2008). As such, this allows a cell-by-cell analysis for quantitation of nuclear, cytoplasmic, and membranous reaction products, applied, for example, for nuclear β-catenin in colon tumors (de Sousa e Melo et al. 2011) and SMAD4 in prostate carcinoma (Ding et al. 2011).
The aim of this study is to develop a method for the reliable determination of the Ki67 proliferation index of hepatocytes with correction for the presence of non-parenchymal cells in the field of interest. We designed IHC double- and triple-staining procedures that were individually analyzed for determining the Ki67 index of hepatocytes only. The application of this procedure may be useful in many other (clinical) settings.
Materials and Methods
Tissue
The tissue samples used for this study are part of another study protocol that will be described in detail elsewhere. The study was approved by the Institutional Animal Ethics Committee of the Academic Medical Center of the University of Amsterdam. Ten New Zealand White rabbits underwent PVE as described elsewhere (van den Esschert et al. 2012). Briefly, cranial liver lobes account for 80% of the total liver volume. Portal vein branches to the cranial lobes were embolized, leading to a variable atrophic response of the cranial lobes and a hyperplastic response of the caudal liver lobe. The animals were sacrificed after 7 days (n=5) or 14 days (n=5). Tissue samples from the left lateral, embolized lobes and the caudal, non-embolized liver lobes were obtained at sacrifice, resulting in 20 tissue blocks in total. Tissue samples were routinely fixed in 4% formalin (48 hr) and processed to paraffin tissue blocks using standard operational procedures. Four-micron sections were cut.
IHC Single, Double, and Triple Staining
Tissue sections were dewaxed in xylene and rehydrated in graded alcohols. Endogenous peroxidase activity was blocked with methanol + 0.3% peroxide (20 min, room temperature). Tissue sections were pretreated with heat-induced antigen retrieval (HIER) performed in a Pretreatment Module (Thermo Fisher Scientific; Fremont, CA) using Tris-EDTA buffer, pH 9.0 (Thermo Fisher Scientific), for 20 min at 98C.
Tris-HCl buffered saline (TBS) was used throughout for washing, and Scytek antibody diluent (Scytek; Logan, UT) was used for diluting reagents. Ultra V Block (Thermo Fisher Scientific) was applied as protein block prior to the first primary antibody. Single IHC staining for Ki67 was performed with Ki67, MIB-5 (Dako, Glostrup, Denmark), visualized by BrightVision horseradish peroxidase (HRP)–conjugated anti-mouse IgG polymer (ImmunoLogic, Duiven, The Netherlands) and BrightDAB. Sections were weakly counterstained with diluted hematoxylin. A sequential IHC triple-staining procedure (van der Loos 2008) started with CD31, clone JC70A (Dako), visualized by BrightVision HRP-conjugated anti-rabbit IgG polymer and BrightDAB (both ImmunoLogic). The DAB reaction product effectively shelters the first set of antibodies and thus prohibits unwanted cross-reactions with later incubation steps (Sternberger and Joseph 1979). Staining was continued with anti-Ki67, MIB-5 (Dako), visualized by BrightVision alkaline phosphatase (AP)–conjugated anti-mouse IgG polymer (ImmunoLogic) and Vector Blue (Vector Laboratories; Burlingame, CA). Next, a HIER step (10 min, 98C) using citrate buffer (pH 6.0) was applied to remove all antibodies used but leaving the chromogens unchanged (van der Loos 2010). Finally, cytokeratin 8+18 (CK), clones K8.8+DC10 (Abcam; Cambridge, UK), was visualized by BrightVision AP-conjugated anti-mouse IgG polymer (ImmunoLogic) and Vector Red (Vector Labs). Slides were weakly counterstained with hematoxylin diluted 1:5 in distilled water for 3 min, washed with tap water, completely dried on a hot plate, and non-aqueous coverslipped with VectaMount (Vector Labs). The Ki67/CK double staining was fully identical to the description above, except that CD31 visualization was skipped.
Multispectral Imaging and Image Analysis
From all IHC double- and triple-stained slides, multispectral data sets of five random fields with Ki67-positive nuclei at ×200 were acquired using a Nuance imaging system (PerkinElmer/Caliper Life Science; Hopkinton, MA) from 420 to 720 nm at intervals of 20 nm. To analyze the frequency of proliferating hepatocytes, MSI data sets were analyzed with Inform 1.4 image analysis software (PerkinElmer/Caliper Life Science) as described by Mansfield (2010). In the typical workflow of the imaging software, the MSI data sets were first spectrally unmixed. Next, the “learn-by-example” interface of the imaging software was used for segmenting the tissue into different categories based on staining and texture (hepatocytes, sinusoidal space, and portal tracts), which were marked by hand drawing (Fig. 2). Segmenting algorithms for IHC double staining and triple staining were developed to correctly segment the tissue categories in all data sets. In double staining, unstained open space between the hepatocytes was used for the learn-by-example interface to recognize sinusoidal space, and CK stained in red supports the interface to recognize hepatocytes. In IHC triple staining, CD31 stained in brown (DAB) enables the learn-by-example interface to recognize sinusoids. Segmentation of hepatocytes and sinusoids was adequate, but bile ducts (also CK positive) in portal tracts were sometimes falsely segmented into the hepatocyte category. If needed, this was hand-corrected. Allowing a cell-by-cell analysis, the hepatocytic and sinusoidal compartments were further segmented down to the cellular level: nuclei (defined by hematoxylin) and cytoplasm (defined by CK, Vector Red).
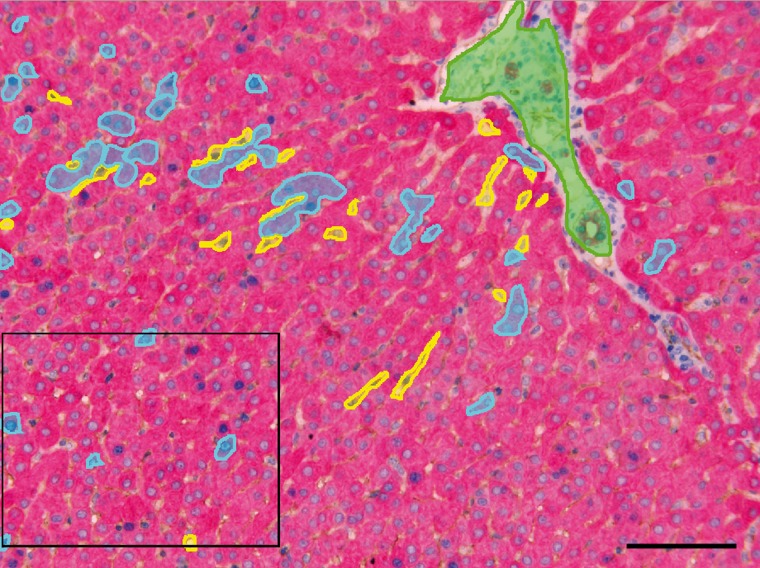
Figure 2.
Rabbit liver tissue section after immunohistochemistry triple staining with cytokeratin (Vector Red), Ki67 (Vector Blue), CD31 (DAB), and hematoxylin counterstaining. Screenshot is taken at the “Segment Tissue” step of the image analysis software. The colored areas in the image are training regions for the software to learn the texture of hepatocytes (cyan), sinusoids (yellow), and portal tract + bile ducts (green). The boxed area is further analyzed in Fig. 3. Bar = 0.1 mm.
At “Score,” the software calculated within the hepatocellular compartment the number of proliferating hepatocytes by analyzing the number of cells with coexpression for cytoplasmic CK (Vector Red) and nuclear Ki67 (Vector Blue). In the sinusoidal compartment, the number of Ki67-positive cells was determined. All spectral data sets were analyzed using the workflow as outlined above and applying similar settings of the software. For comparison and validation of the automated procedure, two observers (CMVDL: observer 1 and JV: observer 2) independently manually counted all Ki67-positive hepatocytes in all RGB images derived from the spectral data sets.
Statistics
Automated and manually counted values were expressed as median (range). Non-parametric tests were used for assessing correlation between measurements (Spearman’s rank coefficient). Correlation coefficients were calculated for the comparison of the manual counting and the hepatocellular Ki67 proliferation index as determined by the cell segmentation inForm software in combination with IHC double or triple staining. Correlation coefficients were also calculated for the manual counting of the two independent observers. A p-value of <0.05 was considered significant. Statistical analysis was performed using SPSS 18 (SPSS, Inc., an IBM Company; Chicago, IL).
Results
Negative controls for IHC single, double, and triple staining replacing the primary antibodies by IgG concentration-matched non-immune reagents were negative for all markers.
Ki67 single staining of the rabbit liver samples resulted in variable amounts of cells with positive nuclear Ki67 staining per slide. However, in most instances, it was unclear on morphological grounds if these cells were proliferating hepatocytes, leucocytes, Kupffer cells, or even other cell types in the field of interest (Fig. 1).
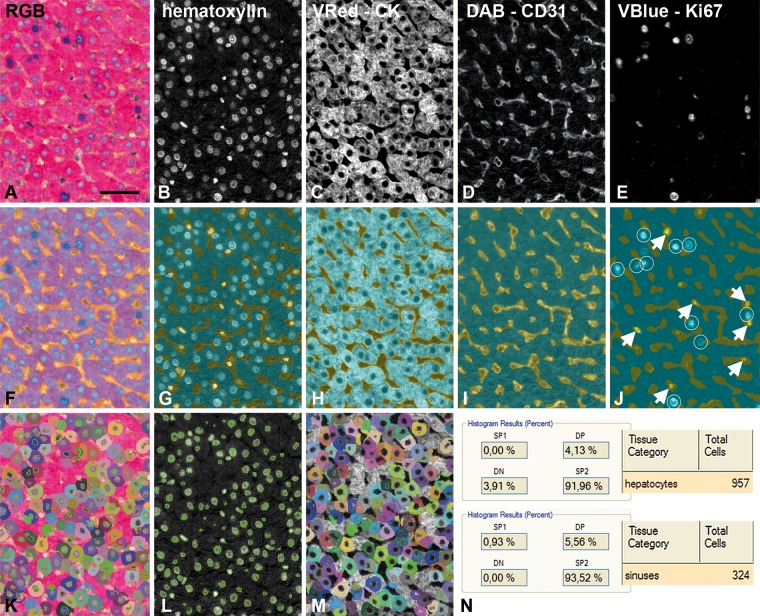
Spectral unmixing resulted in individual images of all chromogen components as shown in Fig. 3A–E for a typical IHC triple-staining experiment. The result of tissue segmentation into hepatocytes, sinusoids, and portal tracts is depicted in Fig. 3F–J. In IHC triple staining, CD31 stained in brown (DAB) enables the learn-by-example interface to recognize sinusoids (Fig. 3I). As a result of tissue segmentation, true Ki67-positive hepatocytes are in the hepatocyte tissue category and non-parenchymal Ki67-positive cells are in the sinusoidal space (Fig. 3J). After cell segmentation (Fig. 3K–M) the total number and the percentage of double positive cells (cytoplasmic CK, Vector Red and nuclear Ki67, Vector Blue) were determined by the software.
Figure 3.
Detail of a rabbit liver tissue section immunohistochemistry (IHC) triple staining in the different stages of automated analysis. (A) RGB image, detail from boxed area in Fig. 1. (B–E) Chromogen components after spectral unmixing: hematoxylin (B); cytokeratin, Vector Red (C); CD31, DAB (D); and Ki67, Vector Blue (E). (F) Tissue segmentation: sinusoids (yellow) and hepatocytes (cyan). (G–J) Tissue segmentation with respect to the different chromogen components. Note that in the Ki67, Vector Blue component (J), Ki67-positive nuclei are partly in the sinusoids marked in yellow (arrows) and partly in the hepatocyte area (white circles). (K) Cell segmentation composed of nuclear segmentation (L, hematoxylin) and cytoplasmic segmentation (M, cytokeratin, Vector Red). (N) Final scoring shows that in the image as in Fig. 2, there are 957 hepatocytes; DP (double positive) = 4.13% of them are Ki67-positive (= 40 cells), SP1 (single positive Ki67, Vector Blue) cells are not present, and DN (double negative) cells = 3.91%. SP2 (single positive cytokeratin, Vector Red) = 91.96 means that the vast majority of hepatocytes are Ki67 negative. In the sinusoids, there are 324 cells, and 5.56% of them are Ki67 positive (= 18 cells). Bar = 0.05 mm.
IHC Double-staining CK and Ki67
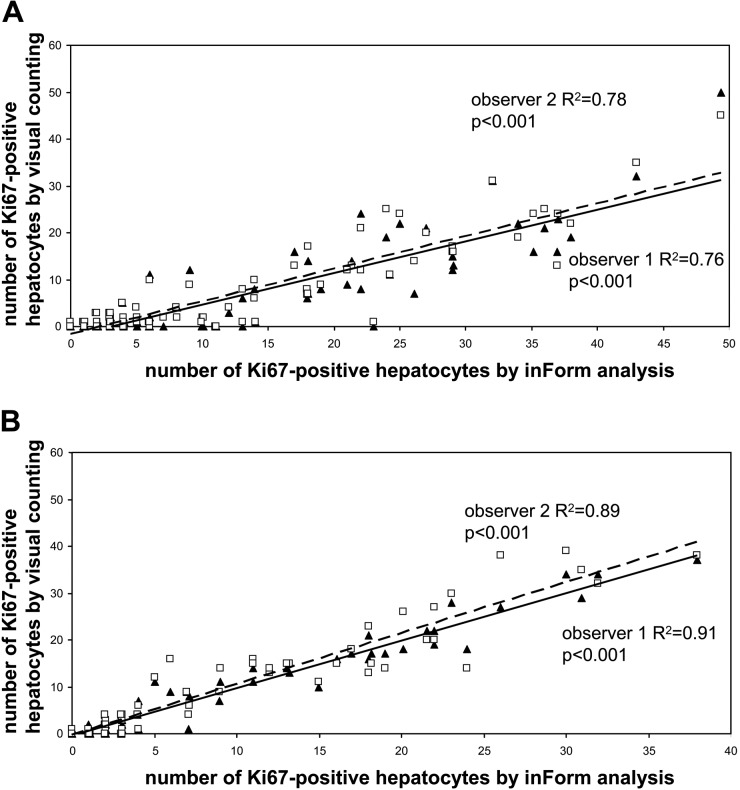
The numbers of Ki67-positive hepatocytes were manually counted by two independent observers using a CK/Ki67 double staining, and the results were compared with the data acquired using the Inform software. The median and range of Ki67-positive hepatocytes manually counted in the IHC double staining by the two observers were 0 (0–50) for observer 1 and 2 (0–45) for observer 2 (R2 = 0.86, p<0.001). The digital analyses using the software counted 6 (0–49) positive nuclei with a good correlation to the manual counting of both observer 1 (R2 = 0.76, p<0.001) and observer 2 (R2 = 0.78, p<0.001) (Fig. 4A). Median and range of Ki67-positive cells in the sinusoids as calculated by the software were 7 (1–40).
Figure 4.
Number of Ki67-positive hepatocytes in rabbit liver tissue sections with automated analysis versus visual counting by two observers in all images analyzed (n=100; i.e., five random fields in 20 tissue blocks at ×200 magnification). (A) Analysis of CK-Ki67 immunohistochemistry (IHC) double staining. (B) Analysis of CD31-CK-Ki67 IHC triple staining. Note the higher correlation coefficients after IHC triple-staining. ▲ = observer 1; □ = observer 2.
IHC Triple-staining CK, CD31, and Ki67
The median, ranges, and correlation coefficients of Ki67-positive hepatocytes manually counted in the IHC triple staining by the two observers were 1 (0–37) for observer 1 and 1 (0–39) for observer 2 (R2 = 0.95, p<0.001). The digital analyses using the software counted 2 (0–38) positive nuclei with higher correlation coefficients to the manual counting than with IHC double staining: observer 1 (R2 = 0.91, p<0.001) and observer 2 (R2 = 0.89, p<0.001) (Fig. 4B). Median and range of Ki67-positive cells in the sinusoids as calculated by the software were 9 (1–20). Total cell counts of Ki67-positive cells in hepatocyte and sinusoid tissue categories by software analysis are shown in Table 1. This table reveals the highly variable amount of Ki67-positive inflammatory cells in the sinusoids, potentially hampering reliable determination of the proliferation index by Ki67 single staining.
Table 1.
Total Counts of Ki67-positive Cells by Automated Analysis of the Immunohistochemistry Triple-Staining Results
| Rabbit Sample | Proliferating Hepatocytes | Proliferating Cells in Sinusoids |
|---|---|---|
| 1 | 3 | 38 |
| 2 | 3 | 37 |
| 3 | 3 | 26 |
| 4 | 12 | 28 |
| 5 | 6 | 60 |
| 6 | 5 | 71 |
| 7 | 5 | 85 |
| 8 | 6 | 72 |
| 9 | 92 | 71 |
| 10 | 2 | 60 |
| 11 | 4 | 49 |
| 12 | 93 | 11 |
| 13 | 65 | 36 |
| 14 | 87 | 40 |
| 15 | 38 | 34 |
| 16 | 3 | 25 |
| 17 | 5 | 58 |
| 18 | 16 | 42 |
| 19 | 151 | 32 |
| 20 | 11 | 42 |
Total cell counts of five fields per rabbit sample are shown in the hepatocellular and sinusoidal compartments. Note that the number of proliferating inflammatory cells in the sinusoidal space heavily varies per rabbit sample and does not correlate with the number of Ki67-positive proliferating hepatocytes.
Discussion
In the present study, we have demonstrated that the hepatocellular Ki67 proliferation index in rabbit liver tissue sections can be reliably and efficiently determined using automated tissue and cell segmentation software in combination with IHC double or triple staining, correcting for other Ki67-positive cell types present in the field of interest. Especially, the IHC triple-staining experiment shows a very high correlation coefficient with manual counting of proliferating hepatocytes.
In our rabbit model, the effects of embolization of the portal vein are studied in different research settings. One of the parameters of interest is the Ki67 hepatocellular proliferation index in the different lobes as a measurement of hypertrophic response of the caudal lobe after selective embolization of the cranial lobes. The outcome of the parameters studied in the present rabbit experiment will be described elsewhere in full detail. We focus here purely on the technical aspects of a novel automated assessment of the Ki67 hepatocellular proliferation index in the different lobes. All slides were pooled together with a wide variability in both influx of inflammatory cells and proliferative activity of the hepatocytes. Applying Ki67 single staining to these rabbit liver specimens hardly allows any visual differentiation between Ki67 staining in hepatocytes and other cell types present in the field of interest, lymphocytes in particular (Fig. 1).
For a more accurate assessment of Ki67 hepatocellular proliferation indices, we originally opted for a double IHC procedure combining Ki67 and CK for marking the hepatocytes. However, it appeared that tissue segmentation into hepatocytes and the sinusoidal compartment was not always ideal due to different sizes and diameters of the sinusoids. As a result, in these narrow and small sinusoids, Ki67-positive leucocytes may be fully surrounded by Vector Red reaction product. As such, this confuses the software algorithms, and consequently, these cells are falsely counted by the software as proliferating hepatocytes, whereas trained observers recognize these cells as leucocytes. An extension of the CK-Ki67 double staining with a leucocyte marker such as leucocyte common antigen (LCA, CD45) would ideally correct for the presence of lymphocytes as the most important cell population interfering with obtaining a reliable measurement of hepatocellular proliferation. However, to our knowledge, such an antibody is not available for formalin-fixed and paraffin-embedded rabbit tissue. Because non-parenchymal Ki67-positive cells, such as leucocytes, Kupffer cells, and endothelial cells, are present in the sinusoids, we tested the approach of marking the liver sinusoids by CD31. This creates an intravascular compartment in which these non-parenchymal cells are seen. Extending the CK-Ki67 double staining with an additional CD31 staining facilitates the software for optimal tissue segmentation into a sinusoidal compartment and a hepatocellular compartment. The improved tissue segmentation obviously excludes the proliferating cells present in the sinusoids (Fig. 3J) and therefore results in a more accurate evaluation of the number of Ki67-positive proliferating hepatocytes. This is reflected in a higher correlation coefficient for the IHC triple staining as compared with the IHC double staining (Fig. 4). Table 1 shows that the actual number of Ki67-positive cells in both the hepatocellular compartment and the sinusoidal compartment in these rabbit tissue samples is extremely variable. This means that counting of all Ki67-positive cells from single staining specimens as seen in Fig. 1 does not yield an accurate number of proliferating hepatocytes. The IHC triple staining, in combination with the software for tissue segmentation into hepatocytes, the sinusoidal compartment, and portal tracts, significantly improved correct evaluation of the actual number of proliferating hepatocytes.
The Ki67 proliferation index is required for grading different tumor types, such as neuroendocrine tumors and neoplasms of the central nervous system (Prayson 2005; Rindi et al. 2007; Preusser et al. 2012; Zahel et al. 2012). These are tumors in which a substantial number of tumor-infiltrating lymphocytes can be found, which hampers reliable determination of the proliferation index in daily practice when single staining for Ki67 is used. The degree of lymphocytic infiltration of tumors may even differ between the primary tumor and metastasis (Vikman et al. 2009). In tumors of the central nervous system, reactive glial cells are found within the tumor together with proliferating endothelial cells, and of course also with inflammatory cells. This may lead to an overestimation of the tumor proliferation index, particularly in low-grade tumors with less predominance of tumor cells (Prayson 2005). Although the technique described in this article still has to be validated in these specific clinical settings, we believe that IHC double or, even better, triple staining, in combination with automated tissue and cell segmentation software, might be very useful in clinical practice for obtaining fast and reliable tumor proliferation indices.
In conclusion, IHC double or triple staining in combination with the multispectral tissue and cell segmentation inForm software reliably determines the proliferative activity of hepatocytes in a rabbit model and has the potential for broad applicability in clinical practice when the Ki67 proliferation index yields prognostic significance.
Footnotes
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
References
- de Graaf W, van den Esschert JW, van Lienden KP, Roelofs JJ, van Gulik TM. 2011. A rabbit model for selective portal vein embolization. J Surg Res. 171:486–494 [DOI] [PubMed] [Google Scholar]
- de Graaf W, van Lienden KP, van den Esschert JW, Bennink RJ, van Gulik TM. 1998. Increase in future remnant liver function after preoperative portal vein embolization. Br J Surg. 98:825–834 [DOI] [PubMed] [Google Scholar]
- de Sousa e Melo, Colak FS, Buikhuisen J, Koster J, Cameron K, de Jong J, Tuynman J, Prasetyanti Fessler PE, van den Bergh S, Rodermond H, et al. 2011. Methylation of cancer stem cell-associated Wnt target genes predicts poor prognosis in colorectal cancer patients. Cell Stem Cell. 9:476–485 [DOI] [PubMed] [Google Scholar]
- Ding Z, Jiun Wu C, Chu GC, Xiao Y, Ho D, Zhang J, Perry SR, Labrot ES, Wu X, Lis R, et al. 2011. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature. 470:269–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson RM. 2008. Putting the more “back” in morphology: spectral imaging and image analysis in the service of pathology. Arch Pathol Lab Med. 133:748–757 [DOI] [PubMed] [Google Scholar]
- Mansfield JR. 2010. Cellular context in epigenetics: quantitative multicolor imaging and automated per-cell analysis of miRNAs and their putative targets. Methods. 52:271–280 [DOI] [PubMed] [Google Scholar]
- Prayson RA. 2005. The utility of MIB-1/Ki-67 immunostaining in the evaluation of central nervous system neoplasms. Adv Anat Pathol. 12:144–148 [DOI] [PubMed] [Google Scholar]
- Preusser M, Hoeftberger R, Woehrer A, Gelpi E, Kouwenhoven M, Kros JM, Sanson M, Idbaih A, Brandes AA, Heinzl H, et al. 2012. Prognostic value of Ki67 index in anaplastic oligodendroglial tumours—a translational study of the European Organization for Research and Treatment of Cancer Brain Tumor Group. Histopathology. 60:885–894 [DOI] [PubMed] [Google Scholar]
- Rindi G, Klöppel G, Couvelard A, Komminoth P, Körner M, Lopes JM, McNicol AM, Nilsson O, Perren A, Scarpa A, et al. 2007. TNM staging of midgut and hindgut (neuro) endocrine tumors: a consensus proposal including a grading system. Virchows Arch. 451:757–762 [DOI] [PubMed] [Google Scholar]
- Sternberger LA, Joseph SA. 1979. The unlabeled antibody method: contrasting color staining of paired pituitary hormones without antibody removal. J Histochem Cytochem. 27:1424–1429 [DOI] [PubMed] [Google Scholar]
- van den Esschert JW, van Lienden KP, Alles LK, van Wijk AC, Heger M, Roelofs JJ, van Gulik TM. 2012. Liver regeneration after portal vein embolization using absorbable and permanent embolization materials in a rabbit model. Ann Surg. 255:311–318 [DOI] [PubMed] [Google Scholar]
- van den Esschert JW, van Lienden KP, de Graaf W, Maas MA, Roelofs JJ, Heger M, van Gulik TM. 2011. Portal vein embolization induces more liver regeneration than portal vein ligation in a standardized rabbit model. Surgery. 149:378–385 [DOI] [PubMed] [Google Scholar]
- van der Loos CM. 2008. Multiple immunoenzyme staining: methods and visualizations for the observation with spectral imaging. J Histochem Cytochem. 56:313–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Loos CM. 2010. Chromogens in multiple immunohistochemical staining used for visual assessment and spectral imaging: the colorful future. J Histotechnol. 33:31–40 [Google Scholar]
- Vikman S, Sommaggio R, De La, Torre M, Oberg K, Essand M, Giandomenico V, Loskog A, Totterman TH. 2009. Midgut carcinoid patients display increased numbers of regulatory T cells in peripheral blood with infiltration into tumor tissue. Acta Oncol. 48:391–400 [DOI] [PubMed] [Google Scholar]
- Zahel T, Krysa S, Herpel E, Stenzinger A, Goeppert B, Schirmacher P, Hoffmann H, Schnabel PA, Warth A. 2012. Phenotyping of pulmonary carcinoids and a Ki67-based grading approach. Virchows Arch. 460:299–308 [DOI] [PubMed] [Google Scholar]