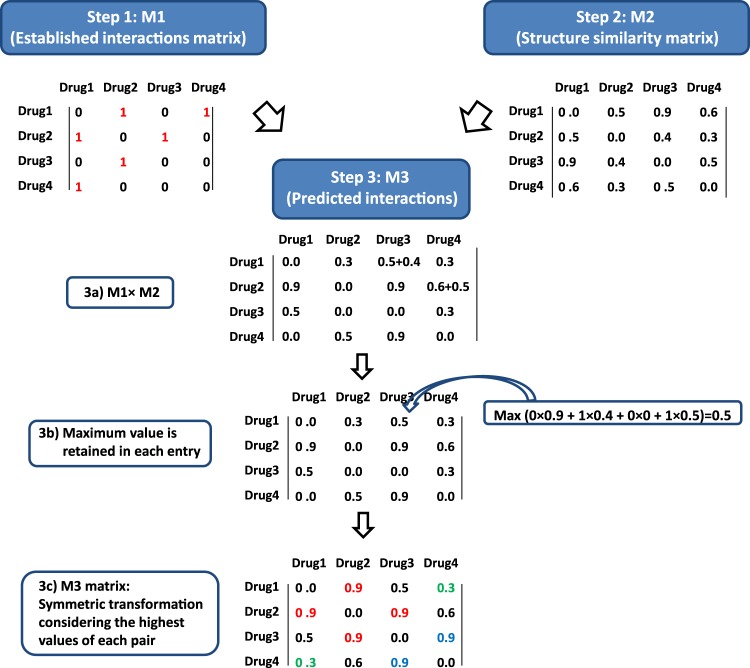
Figure 2.
Generating a drug–drug interactions (DDI) similarity model through combination of the DrugBank interaction database and molecular fingerprint-based modeling. In step 1, interaction matrix M1 is created where the interactions in DrugBank are represented as ‘1’. In step 2, the similarity matrix M2 is created based on the Tanimoto coefficient values. In step 3, M1×M2 is performed, the maximum value for each entry is retained, and the final matrix, M3, is formed based on a symmetry-based transformation (retrieved interactions from M1 when TC>0.75 are represented in red, new predicted interactions with TC>0.75 are represented in blue, and non-retrieved interactions from M1 when TC>0.75 are represented in green). Values in the diagonal of all the matrices are 0 because the interaction of a drug with itself is not considered.