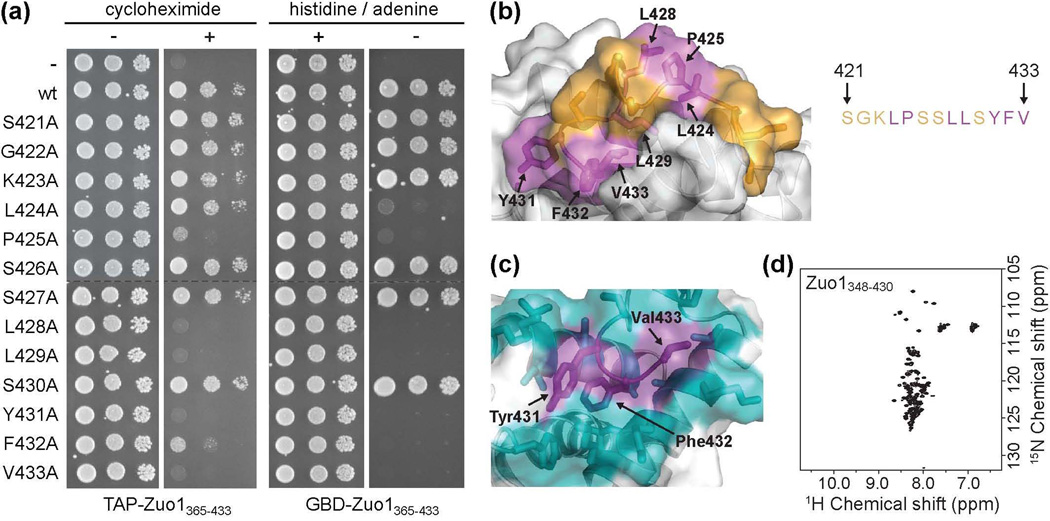
Figure 5.
Residues critical for Pdr1 activation are buried in the C-terminal helical bundle. (a) Alanine scanning mutagenesis of residues 421-433. (left) Wt cells were transformed with vector containing DNA encoding TAP (-) or TAP fused to wt Zuo1365–433 (wt) or Zuo1365–433 containing one of the 13 alanine point mutations and plated onto medium without (−) or with (+) cycloheximide. (right) PJ6933 cells were transformed with vector containing DNA encoding GBD (−) or GBD fused to wt Zuo1365–433 (wt) or Zuo1365–433 containing one of the 13 alanine point mutations and plated onto medium with (+) or without (−) histidine and adenine to detect auto-activation of Gal1-HIS3 and Gal2-ADE2 reporters. All serial dilutions on the same medium were plated on the same plate. Dotted lines indicate where the columns were aligned for display purposes. (b) Residues important for Pdr1 activation, as determined by loss of activity upon alteration to alanine as described in (a), are indicated in magenta. Residues in which substitution to alanine had no effect on activity in either assay are shown in orange. (c) The three most C-terminal residues, Tyr431, Phe432, and Val433 (purple), are well constrained in the hydrophobic core of the domain, as indicated by the large number of NOEs observed between these and surrounding residues, shown in cyan. (d) 15N–1H HSQC spectrum of the C-terminal domain lacking the three most C-terminal residues (Zuo1348-430).