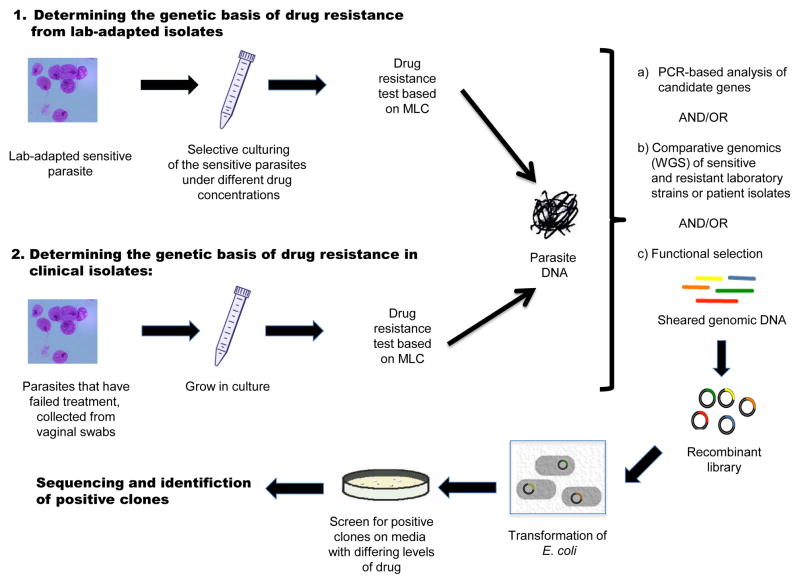
Figure 2. Genomic methods for the characterization of drug resistance.
Two approaches are shown: (1) Selection of drug resistant lines from culture-adapted sensitive strains grown in media containing increasing levels of drug (lab-induced anaerobic resistance); and (2) Isolation of drug resistant isolates from vaginal swabs (clinical aerobic resistance). In both (1) and (2), after quantifiable determination of the phenotype (minimal lethal concentration, or MLC) and DNA extraction, one of three steps or a combination of all can be applied: (a) Analysis of candidate genes known to be involved in drug resistance in other microbes; (b) Whole genome sequencing (WGS) and comparative genomics of sensitive and resistant strains; (c) Functional selection to characterize novel or confirm known resistance genes. In this latter process, genomic DNA is cloned into an expression system vector that can be cultured in the presence of drug. Recombinant clones containing putative resistance genes are able to grow in the presence of drug, and can then be sequenced for identification