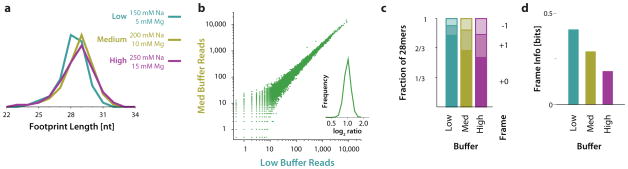
Figure 2.
Buffer conditions affect footprint precision but not expression measurements. (a) Length distribution of ribosome footprints obtained from nuclease digestion in different buffer conditions, as indicated. (b) Expression measurements (average footprint density across each message) by ribosome profiling in different buffer conditions. Each point on the scatter plot represents a human gene, and the expression shown on the two axes is the total number of ribosome footprints in each sample that aligned to the canonical isoform of the gene, excluding those mapping to the first 15 or last 5 codons, where drug treatment distorts ribosome occupancy. The histogram shows the distribution of log2 ratios between medium and low salt/magnesium buffer conditions for genes with at least 200 total footprints in the two measurements. This criterion avoids low-expression genes where statistical sampling error dominates inter-replicate differences. (c) Sub-codon position of ribosome footprint 5′ termini, obtained by nuclease digestion in different buffer conditions, as indicated. Reads that aligned with the annotated CDS of canonical transcripts in the UCSC Known Genes dataset were used, and the position of their 5′ terminus was determined relative to codon boundaries in this CDS. (d) Conditional entropy of the ribosome footprint length and reading-frame position distribution in different samples.