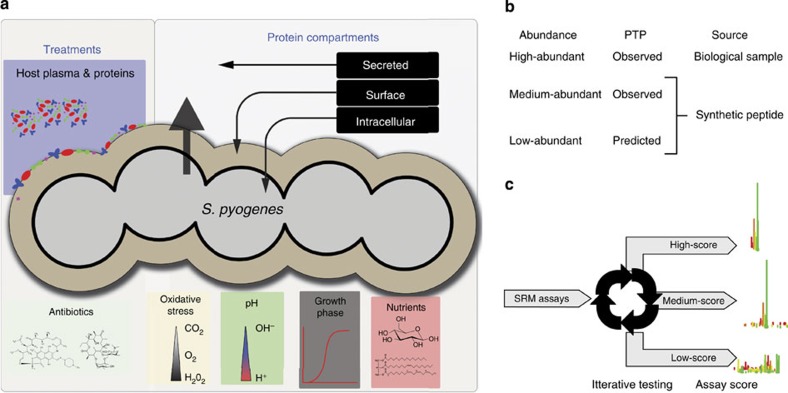
Figure 1. Construction of a proteome-wide SRM assay repository.
(a) Graphical representation of the enriched Group A Streptococci (GAS) cellular compartments. Repeated enrichment of protein pools from the cellular compartments and bacterial states were digested using trypsin and analysed by LC tandem MS (LC–MS/MS). (b) Outline of the strategy used to construct a spectral library from where the low-scoring SRM assays were extracted. For high-abundant PTP’s the SRM assays were determined directly in biological samples, whereas for medium- and low-abundant PTP peptides were synthesized and analysed with LC–MS/MS. (c) To increase the confidence of the individual SRM assays the low-scoring SRM assays were tested extensively in complex mixture of GAS tryptic digest using SRM.