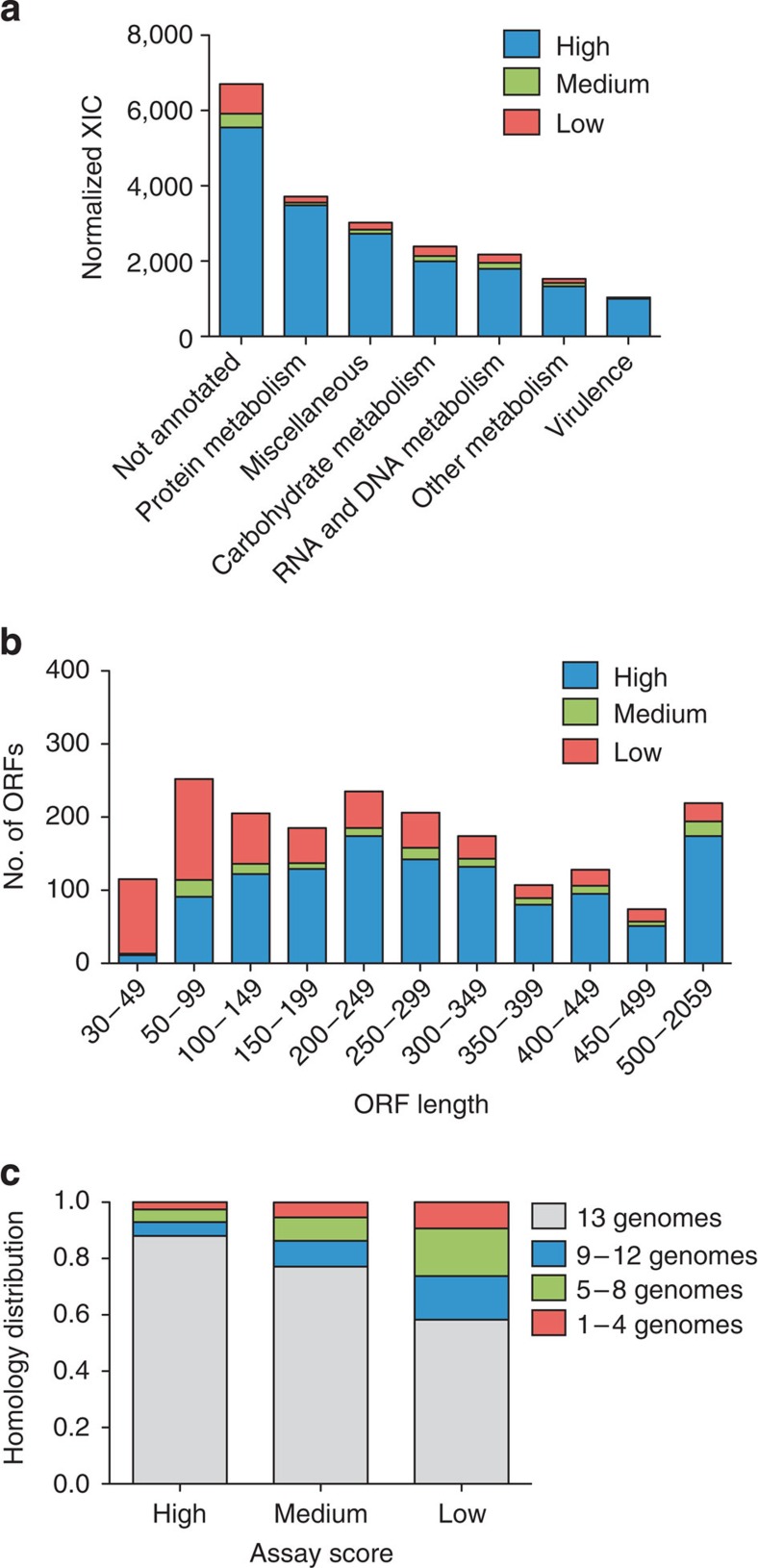
Figure 3. Protein identification biases across functional categories and ORF properties.
Iterative testing of the developed SRM assays in complex biological mixtures of GAS tryptic digests resulted in subdivision of the proteins into three SRM assays-score categories; proteins with low-, medium- or high-scoring SRM assays. Using the SRM assay score categories we determined biases among associated proteins within the three categories. (a) Proportion of XIC intensities associated with proteins with at least one high or medium-scoring SRM assay or proteins with low-scoring SRM assays. NMPDR was used to categorize proteins; protein metabolism includes categories amino acids and derivatives and protein metabolism; miscellaneous includes categories clustering-based sub-systems, miscellaneous, phages, prophages, transposable elements, plasmids, regulation and cell signalling, respiration, stress response, cell division and cell cycle and cell wall and capsule; carbohydrate metabolism includes category carbohydrates; RNA and DNA metabolism includes categories nucleosides and nucleotides, DNA metabolism and RNA metabolism; other metabolism includes categories phosphorus metabolism, potassium metabolism, fatty acids, lipids and isoprenoids, cofactors, vitamins, prosthetic groups and pigments, sulphur metabolism, iron acquisition and metabolism, nitrogen metabolism, membrane transport and metabolism of aromatic compounds virulence includes categories virulence and virulence, disease and defence. (b) Genome-wide correlation between SRM assay score and ORF length. (c) Correlation between SRM assay score and relative degree of protein conservation across 13 GAS strains as determined with TOP-BLAST hits with SF370 as reference.