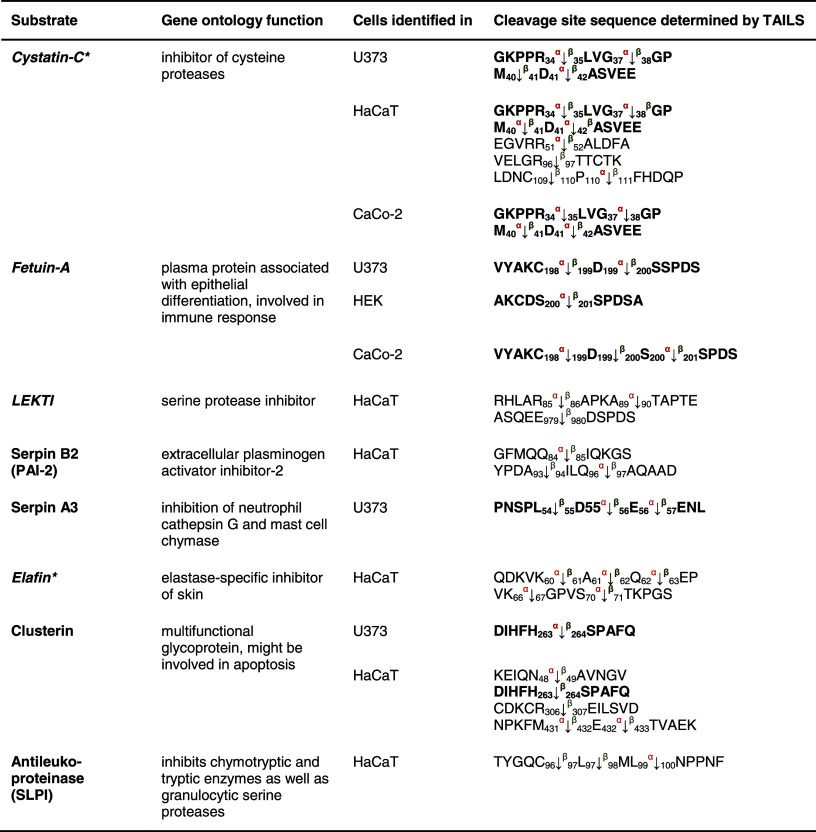
Table 4.
Candidate substrates from the extracellular matrix for meprin α and β identified by TAILS
Protein samples were collected as indicated in Table 1. Except for laminin 5 and the APP, which are known substrates and were also identified by TAILS, the other substrates presented are new candidates for meprin metalloproteases. Proteins in italic have been further validated in vivo (Online resource 3). Sequences are given in the one letter codes. Arrows indicate the cleavage sites analyzed by MS/MS for meprin α (α) and meprin β (β). Numbers display position of amino acid in full-length protein. Confirmed cleavage sites in more than one secretome (biological replicate) are indicated in bold.
TGFBI transforming growth factor-beta-induced protein ig-h3, AD Alzheimer′s disease.