Figure 5.
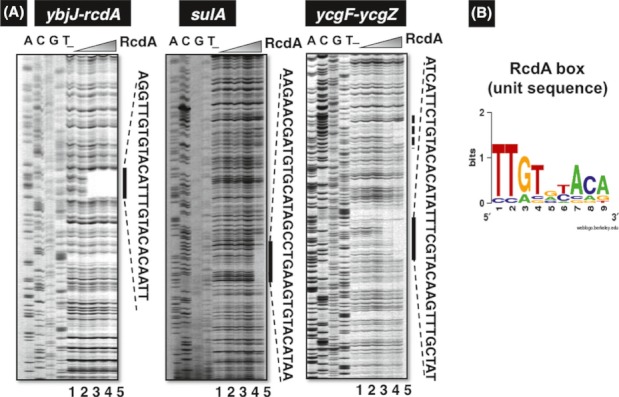
DNase-I footprinting analysis of RcdA-binding sequences. (A) DNase-I footprinting analysis of RcdA-binding sites was performed under the standard conditions (for details see Experimental Procedures) for the ybjJ-rcdA spacer (left panel), sulA ORF (center panel) and ycgF-ycgZ spacer (right panel) sequences. The RcdA concentrations used (left panel) were (from lane 1 to 5): 0 (lane 1), 0.1 (lane 2), 0.5 (lane 3), 1.0 (lane 4), and 5 pmol (lane 5). The sequences protected by RcdA from DNase-I digestion are shown on right side of each panel. The dotted line on left of ycgF-ycgZ fragment (right panel) includes 26-nucleotide-long junction sequence (5′GCTGTCGGAATGGACGA-TTGTATAGA3′) between the vector used for SELEX fragment cloning and the ycgF-ycgZ spacer, of which 5′-proximal 17-nucleotide sequence corresponds to the vector sequence whereas 3′-proximal 9-nucleotide sequence corresponds to ycgF-ycgZ sequence. Noteworthy is that the 3′-proximal sequence contains the RcdA box (TTGTnTACA)-like sequence, which might contribute the generation of footprinting. (B) In addition to these three DNase-I footprinting patterns, the protected sequences of csgD-csgB, ydeI-ydeJ, and ynfM-asr spacer regions by RcdA from DNase-I digestion were used for estimation of the consensus sequence of RcdA binding (data not shown). The logo pattern obtained using WebLogo (http://weblogo.berkeley.edu/logo.cgi) was defined as the RcdA-box unit sequence. The number and location of this unit sequence are different among the RcdA target genes (for details see text).
