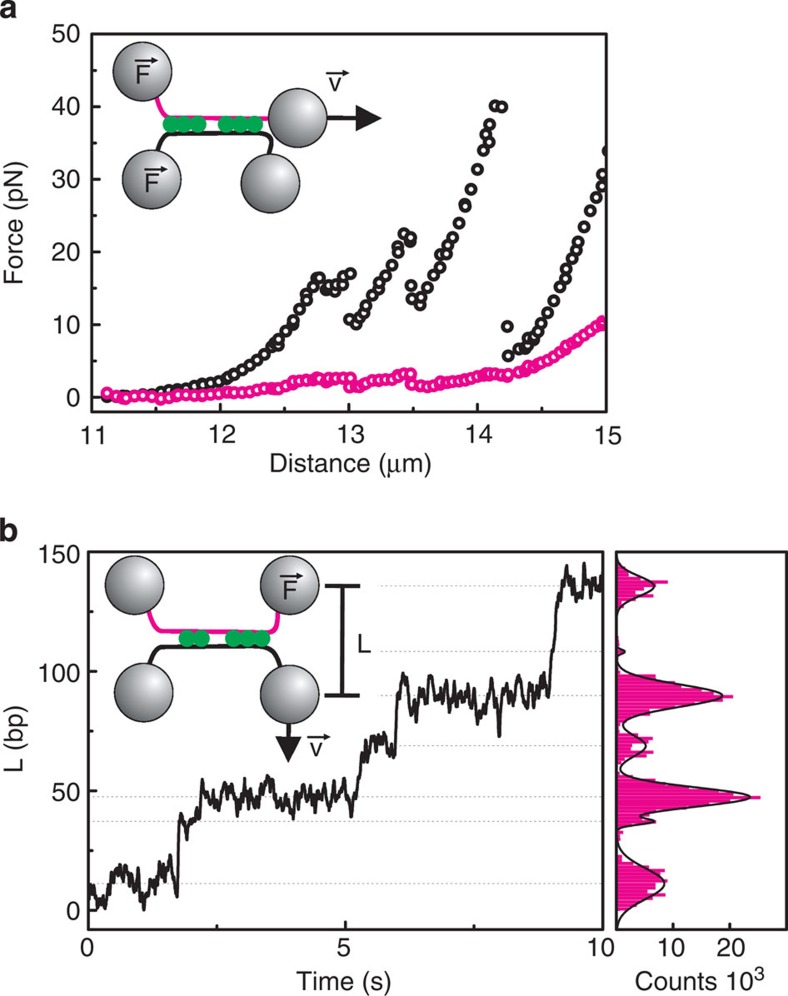
Figure 3. Characterization of Alba-mediated bridges.
Two DNA molecules are crossed and bridged by incubating in 100 nM Alba1 F60A. (a) Shearing experiment. One of the two DNA molecules is stretched with speed v (magenta strand), creating a uniform shearing force (F) over all the formed bridges. The force rises on the opposite molecule (black data points) and relaxes as protein–DNA complexes are ruptured. When the magenta DNA molecule is stretched to ~14.5 μm, the force also rises on the stretched molecule, possibly caused by bridges within the same molecule. (b) Unzipping experiment. A force up to 50 pN is built up over the first bridge between the two DNA molecules, by pulling perpendicular to the DNA molecules with speed v. As protein–DNA complexes are ruptured over time the length of DNA between the two beads (L) becomes larger in discrete steps. The histogram of the time trace is fitted with multiple Gaussians to find the step sizes (right panel), which represent the distance between two adjacent bridges.