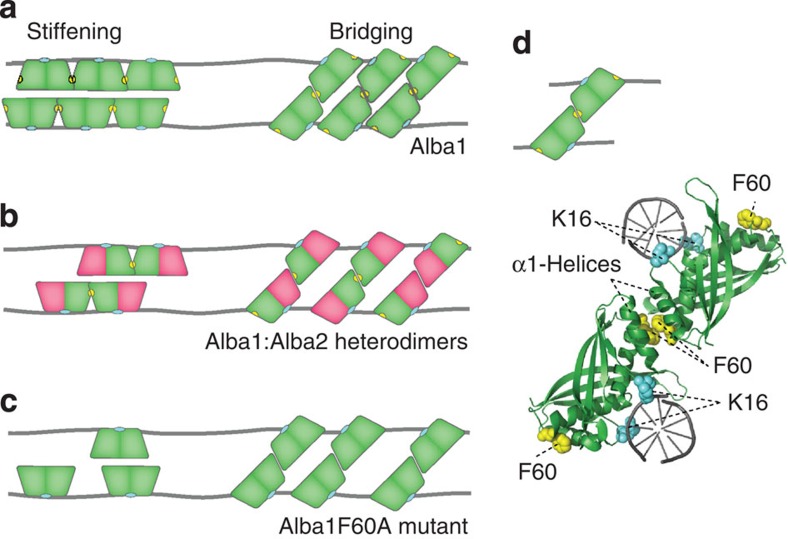
Figure 4. Model of Alba-mediated stiffening and bridging.
(a) Alba1 dimers can bind to the DNA in two different modes depending on their orientation on the DNA (cyan depicts DNA interaction domains). Dimers can either bind side-by-side along the DNA, stiffening the DNA, or interact with dimers on an adjacent duplex, yielding a bridged structure. The F60 residue (yellow) stabilizes dimer–dimer interactions and is responsible for cooperative side-by-side binding. (b) Alba1:Alba2 heterodimers can bind along the DNA but cooperative side-by-side binding is limited to a maximum of two dimers as only the Alba1 subunit contains the F60 residue. Bridged structures can be formed in three different configurations, depending on the orientation of the dimer in the bridged complex. (c) The Alba1 F60A mutant does not exhibit cooperative side-by-side binding along the DNA. Bridges can be formed, but are less stable compared with the Alba1 bridges. (d) Model of Alba1–DNA bridging interaction based on the Alba1 crystal structure (PDB accession code 1H0X22) structurally aligned with the co-crystal structure of (Ape10b2)-dsDNA (PDB accession code 2H9U23) using PyMol. Alba dimers interact with the minor groove of DNA (DNA-interacting K16 residues are shown in cyan). By hydrophobic interactions of the two α-helices and interactions of the F60 residues (yellow) two DNA duplexes are bridged.