Figure 1.
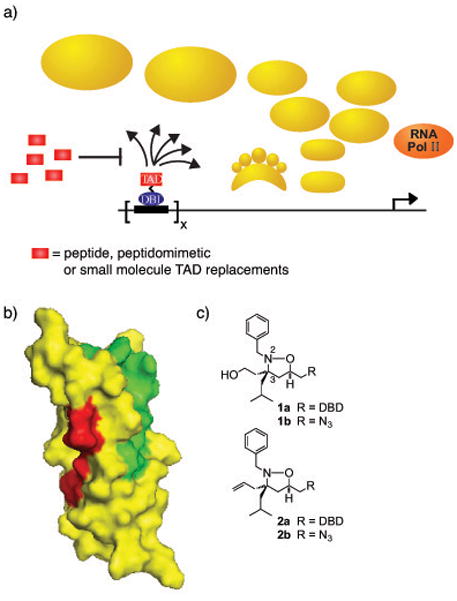
a) Transcriptional activators, minimally comprised of a DNA-binding domain (DBD; blue oval) and a transcriptional activation domain (TAD; red rectangle), upregulate transcription by stimulating chromatin remodeling and facilitating the assembly of the RNA polymerase II holoenzyme at a promoter.14 The TAD is primarily responsible for the direct binding interactions with the transcriptional machinery (gold) in order to accomplish this. Potential binding partners of TADs within the transcriptional machinery include enzymes that modify chromatin, the proteasome, and/or coactivators. Small or large molecule mimics of TADs can serve as competitive inhibitors of activators, thus down-regulating transcription. b) The two activator-binding sites within the KIX domain of CBP/p300. Highlighted in green in the space-filling diagram of the KIX domain are the residues that experience the greatest chemical shift perturbation upon interacting with the TAD of MLL as measured by 1H-15N-HSQC experiments;8b the TADs of Tat, Tax and Jun occupy a similar binding site.8d–f In red are the amino acids that change when interacting with Myb.15 Pymol figures were generated from 1 kdx. c)Isoxazolidine-based mimics of transcriptional activation domains. DBD = OxDex conjugated to an ethylene glycol linker (AEEA).13
