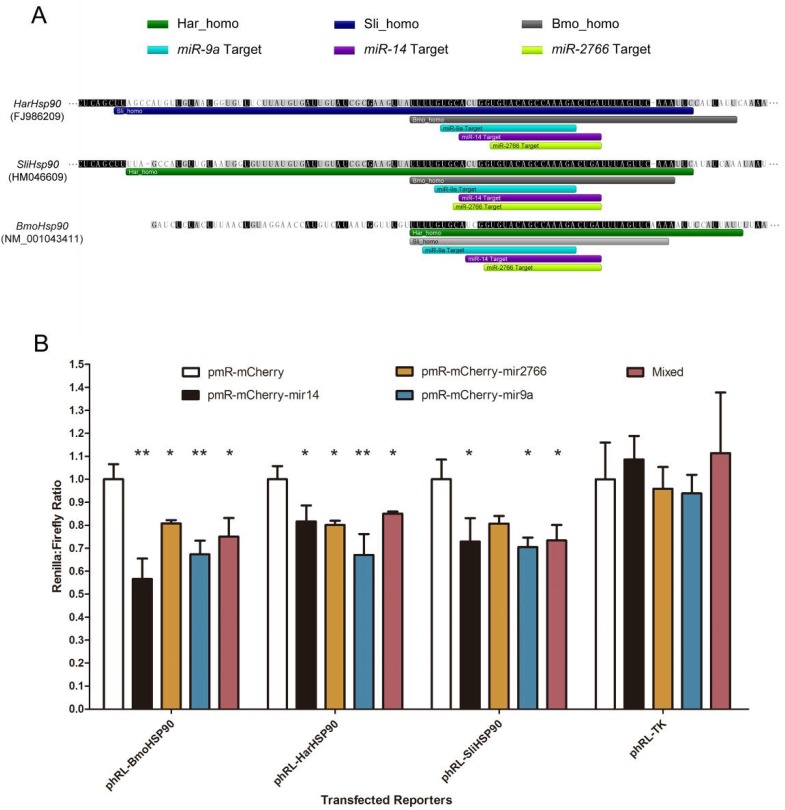
Figure 5.
Conserved miRNA target sites in 3' UTRs of Hsp90 genes. (A) Homologous regions and miRNA target sites in the 3' UTRs of Hsp90 genes. The bars indicate regions homologous to H. armigera (dark green), S. litura (dark blue) or B. mori (grey) and predicted miRNA target sites of miR-9a (light blue), miR-14 (purple) or miR-2766 (light green). (B) Down-regulation of reporter gene expression with the 3' UTRs of Hsp90 genes. miRNA precursors were inserted into pmR-mCherry for miRNA over-expression. These miRNA-expressing plasmids were co-transfected into 293T cells with the pGL3-promoter and pRL-TK parental (Renilla control) or pRL-TK-3'UTR plasmids into which the Hsp90 3' UTRs were respectively cloned. The y-axis shows the Renilla luciferase activity (relative luminescence units [RLU]) normalized to the firefly luciferase activity and compared with the vector control, which was set to 1.0 within each experiment. The values are the average±SD of 3 biological repeats. The abbreviations represent the species as follows: Bmo, B. mori; Har, H. armigera; Sli, S.litura. A paired t-test was used to analyze the significance of the differences. A difference with p-value less than 0.05 was considered significant (marked with "*" in the figure) and less than 0.01 was considered highly significant (marked with "**").