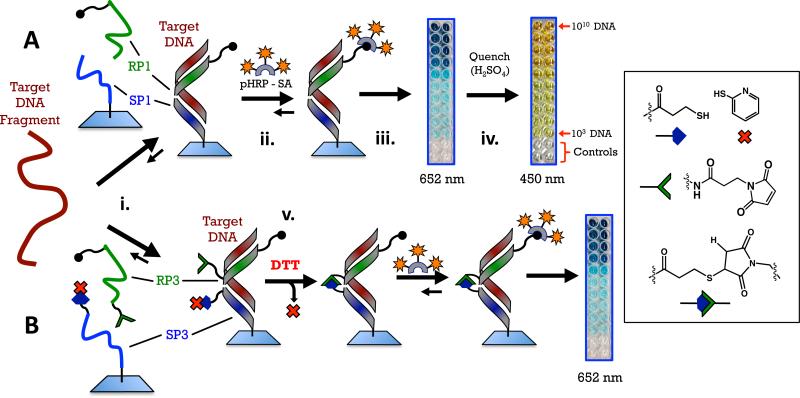
Figure 3.
Schemes for the two detection methods using PNA probes. A. Sandwich: PNA probes RP1 and SP1 recognize a 27-base sequence of anthrax DNA and form a three-component, non-covalent complex on a plastic surface (step i). Probe RP1 has six biotins on the C-terminal. If the target DNA is present and the complex forms, a color can be developed by introducing a polyhorseradish peroxidase+streptavidin conjugate followed by a solution of tetramethylbenzidine and peroxide (step ii). Enzymatic oxidation initially produces a blue color at 652 nm (step iii), quenching the enzyme with sulfuric acid produces a yellow color at 450 nm (step iv). B. Crosslinked sandwich: PNA probes RP3 and SP3 have reactive maleimide and thiol groups on their N- and C-terminal ends, respectively. A disulfide with 2-pyridylthiol is used as a protecting group for the pendant thiol on SP3. This group is reduced with dithiothreitol immediately following hybridization of the PNA probes with DNA to reveal a pendant thiol that reacts with maleamide on the adjacent PNA (step v).