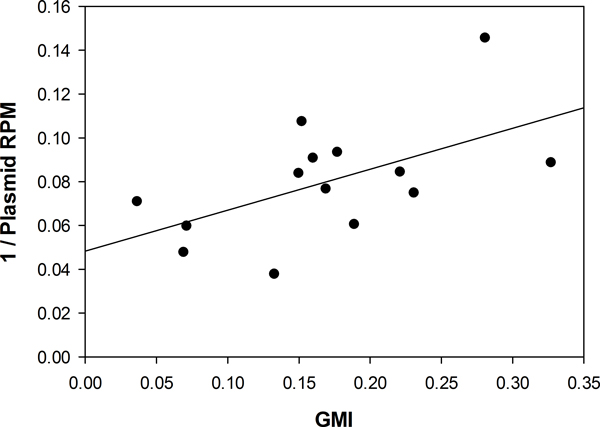
Figure 4.
Global methylation indicator scales inversely with read counts from a spiked-in in vitro methylated construct. The pIRES2-EGFP plasmid was in vitro methylated and spiked-in at a set concentration into each of 14 samples from the decitabine study prior to sequencing. After sequencing, GMI was calculated and plotted against the inverse of the number of normalized reads aligning to the plasmid, and a linear best fit drawn through the points (p = 0.036, R2 = 0.318).