Fig 1.
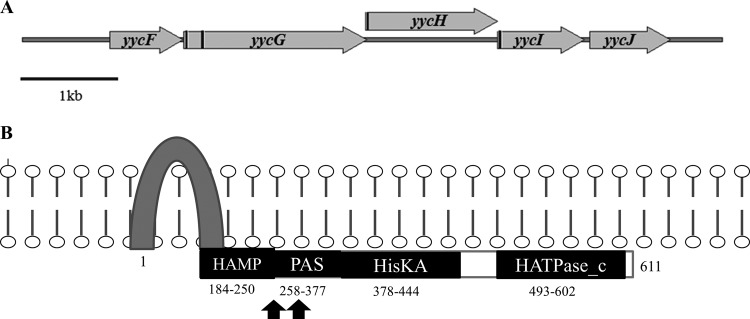
The Yyc system in E. faecium. (A) Structural organization of the orthologous yyc gene cluster in E. faecium; transmembrane coding regions are marked as black bars. Annotations are based on a published report by Türck et al. (49). (B) Predicted domain architecture of the putative YycG histidine kinase and changes found in E. faecium R446, TX0133a, and TX0133a.4, with black arrows denoting the localization of the predicted amino acid changes. Numbers indicate the corresponding amino acids spanning each domain. The HAMP domain is predicted to promote communication between the input and output domains; the PAS sensor domain was named for homology to the PER-ARNT-SIM proteins of D. melanogaster; HisKA, phosphoacceptor domain of histidine kinases; HATPase_c, histidine kinase-like ATPase. The scheme was based in part on published data from Dubrac et al. (56).
