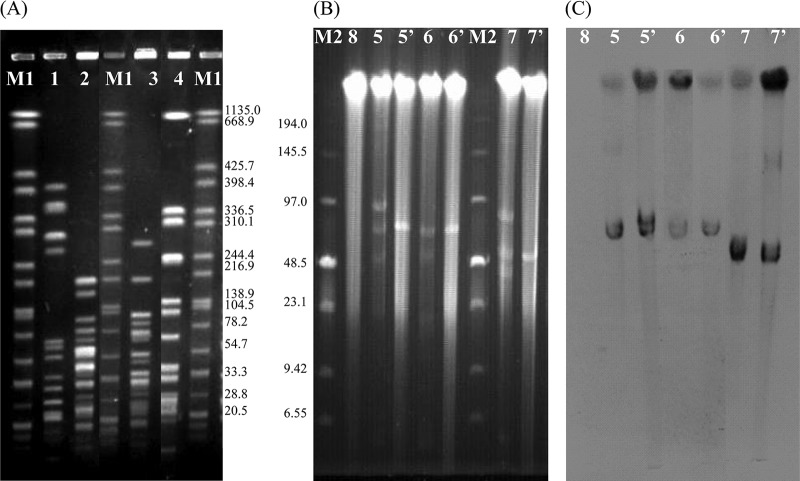
Fig 1.
(A) SmaI-PFGE patterns from E. faecalis EF-01 and three cfr-positive isolates used in the present study. Lanes 1 to 4, E. faecalis EF-01, E. thailandicus W3, E. thailandicus 3-38, and E. faecalis W9-2, respectively; lanes M1 contain the XbaI pattern of Salmonella braenderup H9812 with the fragment sizes given in kilobases on the right-hand side. (B) Numbers and sizes of plasmids as determined by S1 nuclease PFGE. Lane M2, low-range PFG marker (NEB); lanes 5 and 5′, W3R and its transconjugant JHW3; lanes 6 and 6′, 3-38 and its transconjugant JH3-38; lanes 7 and 7′, W9-2 and its transconjugant JHW9-2; lane 8, the recipient E. faecalis JH2-2. (C) Southern hybridization with the cfr probe. The lane numbers correspond to those in panel B.