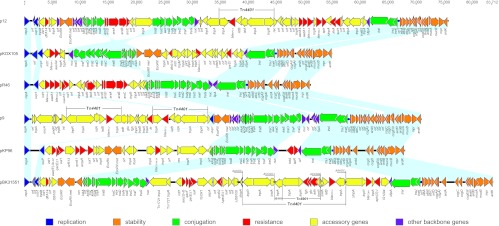
Fig 1.
Comparative analysis of IncN plasmids pBK31551 (JX193301), pKP96 (EU195449), p9 (FJ223607), pR46 (AY046276), pKOX105 (HM126012), and p12 (FJ223605). Light blue shading denotes shared regions of homology. It is noted that the region between kikA and nuc is inverted in pKOX105 relative to the other plasmids and is split into two regions in p12 due to the insertion of the ars operon. Open reading frames (ORFs) are portrayed by arrows and colored based on predicted gene function. Resistance genes are indicated in red arrows and include aminoglycoside resistance genes [aph (3′)-I, aac(3)-Ib, aac(6′)-Ib-cr, aph, aadA1, and aacA4], β-lactamase genes (blaKPC-2, blaKPC-3, blaKPC-4, blaKPC-5, blaVIM-1, blaCTX-M-24, blaOXA-2, blaOXA-9, blaTEM-1, blaSHV-12, and ampR), quinolone resistance genes (qnrA1, qnrB2, and qnrS1), arsenic resistance genes (arsA, arsB, arsC, arsD, and arsR), tetracycline resistance genes (tetA and tetR), and quaternary compound, trimethoprim, and sulfonamide resistance genes (sul1, qacF, qacEΔ1, and dfrA14), as well as glyoxalase/bleomycin resistance gene glo, alcohol dehydrogenase gene adh, and S-formylglutathione hydrolase gene fgh.